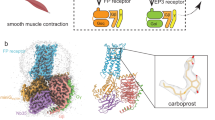
Abstract
Misoprostol is a life-saving drug in many developing countries for women at risk of post-partum hemorrhaging owing to its affordability, stability, ease of administration and clinical efficacy. However, misoprostol lacks receptor and tissue selectivities, and thus its use is accompanied by a number of serious side effects. The development of pharmacological agents combining the advantages of misoprostol with improved selectivity is hindered by the absence of atomic details of misoprostol action in labor induction. Here, we present the 2.5 Å resolution crystal structure of misoprostol free-acid form bound to the myometrium labor-inducing prostaglandin E2 receptor 3 (EP3). The active state structure reveals a completely enclosed binding pocket containing a structured water molecule that coordinates misoprostol's ring structure. Modeling of selective agonists in the EP3 structure reveals rationales for selectivity. These findings will provide the basis for the next generation of uterotonic drugs that will be suitable for administration in low resource settings.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The misoprostol-FA EP3 receptor complex structure coordinates and structure factors are available via the Protein Data Bank (PDB) accession code 6M9T.
Change history
20 December 2018
In the version of this article originally published, the present address for Petr Popov was incorrectly listed as ‘Koltech Institute of Science & Technology, Moscow, Russia’. The correct present address is ‘Skolkovo Institute of Science and Technology, Moscow, Russia’. The error has been corrected in the HTML and PDF versions of the paper.
References
Ricciotti, E. & FitzGerald, G. A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31, 986–1000 (2011).
Woodward, D. F., Jones, R. L. & Narumiya, S. International Union of Basic and Clinical Pharmacology. LXXXIII: classification of prostanoid receptors, updating 15 years of progress. Pharmacol. Rev. 63, 471–538 (2011).
Wang, D. & Dubois, R. N. Eicosanoids and cancer. Nat. Rev. Cancer 10, 181–193 (2010).
Michelson, A. D. Antiplatelet therapies for the treatment of cardiovascular disease. Nat. Rev. Drug. Discov. 9, 154–169 (2010).
O'Callaghan, G. & Houston, A. Prostaglandin E2 and the EP receptors in malignancy: possible therapeutic targets? Br. J. Pharmacol. 172, 5239–5250 (2015).
Hao, C.-M. & Breyer, M. D. Physiological regulation of prostaglandins in the kidney. Annu. Rev. Physiol. 70, 357–377 (2008).
Markovič, T., Jakopin, Ž., Dolenc, M. S. & Mlinarič-Raščan, I. Structural features of subtype-selective EP receptor modulators. Drug Discov. Today 22, 57–71 (2017).
Arulkumaran, S. et al. The roles of prostaglandin EP 1 and 3 receptors in the control of human myometrial contractility. J. Clin. Endocrinol. Metab. 97, 489–498 (2012).
Kandola, M. K. et al. EP2 receptor activates dual G protein signaling pathways that mediate contrasting proinflammatory and relaxatory responses in term pregnant human myometrium. Endocrinology 155, 605–617 (2014).
Potts, M., Prata, N. & Sahin-Hodoglugil, N. N. Maternal mortality: one death every 7 min. Lancet 375, 1762–1763 (2010).
Widmer, M. et al. Misoprostol as an adjunct to standard uterotonics for treatment of post-partum haemorrhage: a multicentre, double-blind randomised trial. Lancet 375, 1808–1813 (2010).
Allen, R. & O’Brien, B. M. Uses of misoprostol in obstetrics and gynecology. Rev. Obstet. Gynecol. 2, 159–168 (2009).
Tsai, B. S., Kessler, L. K., Stolzenbach, J., Schoenhard, G. & Bauer, R. F. Expression of gastric antisecretory and prostaglandin E receptor binding activity of misoprostol by misoprostol free acid. Dig. Dis. Sci. 36, 588–593 (1991).
Orobaton, N. et al. Implementing at-scale, community-based distribution of misoprostol tablets to mothers in the third stage of labor for the prevention of postpartum haemorrhage in Sokoto State, Nigeria: early results and lessons learned. PLoS One 12, e0170739 (2017).
Xiang, J. et al. Successful strategies to determine high-resolution structures of GPCRs. Trends Pharmacol. Sci. 37, 1055–1069 (2016).
Ngo, T. et al. Identifying ligands at orphan GPCRs: current status using structure-based approaches. Br. J. Pharmacol. 173, 2934–2951 (2016).
Ballesteros, J. A. & Weinstein, H. in Methods in Neurosciences. Vol. 25 (ed. Sealfon, S.C.) 366–428 (Academic Press, 1995).
Schmid, A., Thierauch, K. H., Schleuning, W. D. & Dinter, H. Splice variants of the human EP3 receptor for prostaglandin E2. Eur. J. Biochem. 228, 23–30 (1995).
Hua, T. et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 547, 468–471 (2017).
Hua, T. et al. Crystal structure of the human cannabinoid receptor CB1. Cell 167, 750–762.e14 (2016).
Audoly, L. & Breyer, R. M. The second extracellular loop of the prostaglandin EP3 receptor is an essential determinant of ligand selectivity. J. Biol. Chem. 272, 13475–13478 (1997).
Audoly, L. & Breyer, R. M. Substitution of charged amino acid residues in transmembrane regions 6 and 7 affect ligand binding and signal transduction of the prostaglandin EP3 receptor. Mol. Pharmacol. 51, 61–68 (1997).
Ungrin, M. D. et al. Key structural features of prostaglandin E2 and prostanoid analogs involved in binding and activation of the human EP1 prostanoid receptor. Mol. Pharmacol. 59, 1446–1456 (2001).
Hanson, M. A. et al. Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012).
Akasaka, H. et al. The key residue within the second extracellular loop of human EP3 involved in selectively turning down PGE2- and retaining PGE1-mediated signaling in live cells. Arch. Biochem. Biophys. 616, 20–29 (2017).
Abramovitz, M. et al. The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim. Biophys. Acta 1483, 285–293 (2000).
Kiriyama, M. et al. Ligand binding specificities of the eight types and subtypes of the mouse prostanoid receptors expressed in Chinese hamster ovary cells. Br. J. Pharmacol. 122, 217–224 (1997).
Negishi, M. et al. TEI-3356, a highly selective agonist for the prostaglandin EP3 receptor. Prostaglandins 48, 275–283 (1994).
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014).
Negishi, M. et al. Functional interaction of prostaglandin E receptor EP3 subtype with guanine nucleotide-binding proteins, showing low-affinity ligand binding. Biochim. Biophys. Acta 1175, 343–350 (1993).
Hamon, M. et al. Modulation of human myometrial PGE2 receptor by GTP characterization of receptor subtype. Prostaglandins 46, 251–268 (1993).
Tsai, B. S., Kessler, L. K., Schoenhard, G., Collins, P. W. & Bauer, R. F. Demonstration of specific E-type prostaglandin receptors using enriched preparations of canine parietal cells and [3H]misoprostol free acid. Am. J. Med. 83, 9–14 (1987).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125 (2018).
Goupil, E. et al. A novel biased allosteric compound inhibitor of parturition selectively impedes the prostaglandin F2alpha-mediated Rho/ROCK signaling pathway. J. Biol. Chem. 285, 25624–25636 (2010).
Popov, P. et al. Computational design of thermostabilizing point mutations for G protein-coupled receptors. eLife 7, e34729 (2018).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Liu, W., Ishchenko, A. & Cherezov, V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 9, 2123–2134 (2014).
Weierstall, U. et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309 (2014).
Barty, A. et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 47, 1118–1131 (2014).
White, T. A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341 (2012).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 68, 352–367 (2012).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D. Biol. Crystallogr. 68, 368–380 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Sawaya, M. R. et al. Protein crystal structure obtained at 2.9 Å resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proc. Natl. Acad. Sci. USA 111, 12769–12774 (2014).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
PyMOL: The PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, LLC., 2015).
Chen, X. et al. Identification of inhibitors of the antibiotic-resistance target New Delhi metallo-β-lactamase 1 by both nanoelectrospray ionization mass spectrometry and ultrafiltration liquid chromatography/mass spectrometry approaches. Anal. Chem. 85, 7957–7965 (2013).
Acknowledgements
This research was supported by NIH R35 GM127086 (V.C.), R21 DA042298 (W.L.), R01 GM124152 (W.L.), the STC Program of the National Science Foundation through BioXFEL (No. 1231306) (U.W. and W.L.), the Russian Science Foundation (project no. 16-14-10273), and the GPCR Consortium. M.A. was supported by a Canadian Institute of Health and Research (CIHR) Postdoctoral Fellowship Award. C.G. acknowledges the Panofsky Fellowship from SLAC National Accelerator Laboratory and Stanford University for financial support. P.P. and V.K. acknowledge the Russian Foundation for Basic Research (RFBR No.18-34-00990). Parts of this research were carried out at the LCLS, a National User Facility operated by Stanford University on behalf of the US Department of Energy and is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC0276SF00515, and at the GM/CA CAT of the Argonne Photon Source, Argonne National Laboratory. We thank A. Walker for assistance in manuscript preparation; M. Chu, K. Villiers and C. Hanson for baculovirus expression and mammalian cell culture, and N. Sawyer for helpful suggestions. We are grateful to F. Badeaux and E. Audet-Badeaux for their encouragement and support.
Author information
Authors and Affiliations
Contributions
M.A. designed the study, crystallized the EP3 receptor, prepared samples for data collection, collected the data for XFEL and synchrotron, and solved and refined the structure. K.L.W. and M.A. performed the binding assays. B.B. performed the signaling assays. B.Z. and V.K. performed ligand docking and selectivity analysis. Y.L. and W.S. performed mass spectrometry. J.V. and D.M. performed molecular biology. P.P. suggested the mutation. C.G. processed the crystallographic XFEL data. A.B. helped with the XFEL data collection. W.L. helped with sample preparation for XFEL data collection. H.H. and U.W. operated the sample injector during XFEL data collection. G.W.H. solved and refined the structure. V.C. supervised XFEL data collection and processing. M.A.H., V.K., and R.C.S. supervised the project. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
B.B is an employee of Domain Therapeutics NA, a company focused on GPCR drug discovery that sells and licenses the cell signaling kit used in this study. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Supplementary Figures 1–8
Rights and permissions
About this article
Cite this article
Audet, M., White, K.L., Breton, B. et al. Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor. Nat Chem Biol 15, 11–17 (2019). https://doi.org/10.1038/s41589-018-0160-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0160-y
This article is cited by
-
Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain
Nature Communications (2023)
-
Docking for EP4R antagonists active against inflammatory pain
Nature Communications (2023)
-
Structures of human prostaglandin F2α receptor reveal the mechanism of ligand and G protein selectivity
Nature Communications (2023)
-
Serial femtosecond crystallography
Nature Reviews Methods Primers (2022)
-
Small-scale approach for precrystallization screening in GPCR X-ray crystallography
Nature Protocols (2020)