Abstract
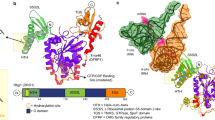
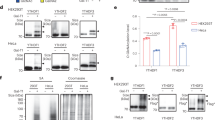
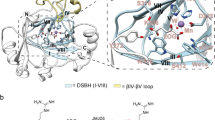
Biochemical, structural and cellular studies reveal Jumonji-C (JmjC) domain-containing 7 (JMJD7) to be a 2-oxoglutarate (2OG)-dependent oxygenase that catalyzes (3S)-lysyl hydroxylation. Crystallographic analyses reveal JMJD7 to be more closely related to the JmjC hydroxylases than to the JmjC demethylases. Biophysical and mutation studies show that JMJD7 has a unique dimerization mode, with interactions between monomers involving both N- and C-terminal regions and disulfide bond formation. A proteomic approach identifies two related members of the translation factor (TRAFAC) family of GTPases, developmentally regulated GTP-binding proteins 1 and 2 (DRG1/2), as activity-dependent JMJD7 interactors. Mass spectrometric analyses demonstrate that JMJD7 catalyzes Fe(ii)- and 2OG-dependent hydroxylation of a highly conserved lysine residue in DRG1/2; amino-acid analyses reveal that JMJD7 catalyzes (3S)-lysyl hydroxylation. The functional assignment of JMJD7 will enable future studies to define the role of DRG hydroxylation in cell growth and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
27 June 2018
In the version of this article initially published, authors Sarah E. Wilkins, Charlotte D. Eaton, Martine I. Abboud and Maximiliano J. Katz were incorrectly included in the equal contributions footnote in the affiliations list. Footnote number seven linking to the equal contributions statement should be present only for Suzana Markolovic and Qinqin Zhuang, and the statement should read “These authors contributed equally: Suzana Markolovic, Qinqin Zhuang.” The error has been corrected in the HTML and PDF versions of the article.
References
Markolovic, S., Wilkins, S. E. & Schofield, C. J. Protein hydroxylation catalyzed by 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 290, 20712–20722 (2015).
Ploumakis, A. & Coleman, M. L. OH, the places you’ll go! Hydroxylation, gene expression, and cancer. Mol. Cell 58, 729–741 (2015).
Semenza, G. L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 9, 47–71 (2014).
Elkins, J. M. et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278, 1802–1806 (2003).
Walport, L. J., Hopkinson, R. J. & Schofield, C. J. Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534 (2012).
Johansson, C. et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics 6, 89–120 (2014).
Morera, L., Lübbert, M. & Jung, M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics 8, 57 (2016).
Chan, M. C., Holt-Martyn, J. P., Schofield, C. J. & Ratcliffe, P. J. Pharmacological targeting of the HIF hydroxylases – A new field in medicine development. Mol. Aspects Med. 47–48, 54–75 (2016).
Cockman, M. E., Webb, J. D. & Ratcliffe, P. J. FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Ann. NY Acad. Sci. 1177, 9–18 (2009).
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009).
Chowdhury, R. et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 510, 422–426 (2014).
Ge, W. et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat. Chem. Biol. 8, 960–962 (2012).
Kato, M. et al. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 39, 1576–1585 (2011).
Feng, T. et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell 53, 645–654 (2014).
Zhuang, Q., Feng, T. & Coleman, M. L. Modifying the maker: Oxygenases target ribosome biology. Translation 3, e1009331 (2015).
Del Rizzo, P. A., Krishnan, S. & Trievel, R. C. Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol. Cell. Biol. 32, 4044–4052 (2012).
Markolovic, S. et al. Structure–function relationships of human JmjC oxygenases—demethylases versus hydroxylases. Curr. Opin. Struct. Biol. 41, 62–72 (2016).
Rose, N. R., McDonough, M. A., King, O. N. F., Kawamura, A. & Schofield, C. J. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 40, 4364–4397 (2011).
Kandoth, C. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Matsunami, N. et al. Identification of rare DNA sequence variants in high-risk autism families and their prevalence in a large case/control population. Mol. Autism 5, 5 (2014).
Lancaster, D. E. et al. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem. J. 383, 429–437 (2004).
Ishikawa, K., Azuma, S., Ikawa, S., Semba, K. & Inoue, J. I. Identification of DRG family regulatory proteins (DFRPs): Specific regulation of DRG1 and DRG2. Genes Cells 10, 139–150 (2005).
Francis, S. M., Gas, M.-E., Daugeron, M.-C., Bravo, J. & Seraphin, B. Rbg1-Tma46 dimer structure reveals new functional domains and their role in polysome recruitment. Nucleic Acids Res. 40, 11100–11114 (2012).
Mantri, M. et al. The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. ChemBioChem 12, 531–534 (2011).
Witkop, B. The application of Hudson’s lactone rule to γ- and δ-hydroxyamino acids and the question of the configuration of δ-hydroxy-L-lysine from collagen. Experientia XII, 372–374 (1956).
Myllyharju, J. & Kivirikko, K. I. Collagens and collagen-related diseases. Ann. Med. 33, 7–21 (2001).
Leśniak, R. K., Markolovic, S., Tars, K. & Schofield, C. J. Human carnitine biosynthesis proceeds via (2S,3S)-3-hydroxy-N ε-trimethyllysine. Chem. Commun. 53, 440–442 (2017).
Daugeron, M.-C., Prouteau, M., Lacroute, F. & Séraphin, B. The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1. Nucleic Acids Res. 39, 2221–2233 (2011).
Ishikawa, K. et al. Cloning and characterization of Xenopus laevis drg2, a member of the developmentally regulated GTP-binding protein subfamily. Gene 322, 105–112 (2003).
Yang, M. et al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 278, 1086–1097 (2011).
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006).
Walport, L. J. et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 (2016).
Mantri, M., Zhang, Z., McDonough, M. A. & Schofield, C. J. Autocatalysed oxidative modifications to 2-oxoglutarate dependent oxygenases. FEBS J. 279, 1563–1575 (2012).
Matheisl, S., Berninghausen, O., Becker, T. & Beckmann, R. Structure of a human translation termination complex. Nucleic Acids Res. 43, 8615–8626 (2015).
Brown, A., Shao, S., Murray, J., Hegde, R. S. & Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015).
Powers, J. C., Asgian, J. L., Ekici, O. D. & James, K. E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 102, 4639–4750 (2002).
Nelson, B. J., Maas, K. J., Dekeyser, J. L. & Stafstrom, J. P. Association of DRG1 and DRG2 with ribosomes from pea, Arabidopsis, and yeast. Int. J. Plant Sci. 170, 834–844 (2009).
Ishikawa, K., Akiyama, T., Ito, K., Semba, K. & Inoue, J. Independent stabilizations of polysomal Drg1/Dfrp1 complex and non-polysomal Drg2/Dfrp2 complex in mammalian cells. Biochem. Biophys. Res. Commun. 390, 552–556 (2009).
Leipe, D. D., Wolf, Y. I., Koonin, E. V. & Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 (2002).
Scotti, J. S. et al. Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc. Natl Acad. Sci. USA 111, 13331–13336 (2014).
Christopher J. Schofield, C. J. & Hausinger, R. P. (eds.). 2-Oxoglutarate-Dependent Oxygenases RSC Metallobiology Series No. 3 (Royal Society of Chemistry, Cambridge, UK, 2015).
Jang, S. H., Kim, A.-R., Park, N.-H., Park, J. W. & Han, I.-S. DRG2 regulates G2/M progression via the cyclin B1-Cdk1 complex. Mol. Cells 39, 699–704 (2016).
Lu, L., Lv, Y., Dong, J., Hu, S. & Peng, R. DRG1 is a potential oncogene in lung adenocarcinoma and promotes tumor progression via spindle checkpoint signaling regulation. Oncotarget 7, 72795–72806 (2016).
Wei, D. et al. Molecular cloning and expression of two closely related GTP-binding proteins from zebrafish. DNA Seq. 15, 246–250 (2004).
Vlangos, C. N., Das, P., Patel, P. I. & Elsea, S. H. Assignment of developmentally regulated GTP-binding protein (DRG2) to human chromosome band 17p11.2 with somatic cell hybrids and localization to the Smith–Magenis syndrome critical interval. Cytogenet. Cell Genet. 88, 283–285 (2000).
de Krom, M. et al. A common variant in DRD3 receptor is associated with autism spectrum disorder. Biol. Psychiatry 65, 625–630 (2009).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Katz, M. J. et al. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc. Natl Acad. Sci. USA 111, 4025–4030 (2014).
Fischer, R. et al. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol. Cell. Proteomics 11, M111.013904 (2012).
Xu, D. et al. Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol. Cell. Proteomics 7, 2215–2228 (2008).
Pérez-Arellano, I., Spínola-Amilibia, M. & Bravo, J. Human Drg1 is a potassium-dependent GTPase enhanced by Lerepo4. FEBS J. 280, 3647–57 (2013).
Gault, J. et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13, 333–336 (2016).
Parsons, T. B. et al. Optimal synthetic glycosylation of a therapeutic antibody. Angew. Chemie Int. Ed. 55, (2361–2367 (2016).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012).
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Acknowledgements
We thank the Biotechnology and Biological Sciences Research Council (BB/L009846/1, C.J.S.), the Medical Research Council (MR/N021053/1, M.L.C.), the Wellcome Trust (106244/Z/14/Z, C.J.S.), Cancer Research UK (24552, M.L.C.), the University of Oxford Clarendon Fund (S.M.) and Kellogg College, University of Oxford (through a Junior Research Fellowship; M.I.A.) for support. We thank E. Flashman (Department of Chemistry, University of Oxford) for assistance with the 18O2 experiment and K. Connolly (Institute of Cancer and Genomics Sciences, University of Birmingham) for Drosophila cDNA.
Author information
Authors and Affiliations
Contributions
The following 'second authors' contributed equally: S.E.W., C.D.E., M.I.A. and M.J.K. S.M., Q.Z., S.E.W., M.J.K., P.W., R.C., M.L.C. and C.J.S. designed and conceived the research; S.M. prepared all recombinant JMJD7 constructs and performed and analyzed CD experiments and all enzyme activity assays by MALDI-MS and NMR; Q.Z. performed substrate discovery proteomics, enzyme–substrate interaction assays, all JMJD7 loss-of-function experiments, and DRG–DFRP interaction, GTPase and RNA-binding assays; S.M. and R.C. performed the crystallography in which R.C. played both an experimental and supervisory role; C.D.E. performed confocal microscopy, cellular dimerization, and dmJMJD7 interaction experiments; Q.Z., C.H. and H.E.M. prepared mammalian expression constructs used by Q.Z.; W.G. and S.M. prepared plasmids for recombinant protein production; S.M. performed and analyzed amino-acid analyses with guidance from S.E.W.; M.I.A. performed and analyzed peptide NMR work, produced and purified recombinant DRG1, performed DRG1 stability and activity assays, and analyzed data; Q.Z. purified exogenous and endogenous proteins for MS and analyzed data with help from M.L.C.; R.K., S.D. and R.F. performed MS and analyzed data; B.M.K. provided MS instrument use. R.K.L. prepared hydroxylysine standards; S.M. and W.B.S. performed and analyzed native MS/SEC-MALS; J.L.P.B. provided SEC-MALS instrument use. M.J.K. and P.W. performed and analyzed Drosophila experiments; R.C. performed enzyme–substrate modeling, prepared recombinant dmJMJD7 and performed proteolysis assays; M.Y., M.E.C. and P.J.R. performed preliminary immunoprecipitations; S.M., Q.Z., S.E.W., C.D.E., R.C., M.L.C. and C.J.S. analyzed data; S.M., Q.Z., R.C., M.L.C. and C.J.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3 and Supplementary Figures 1–39
Rights and permissions
About this article
Cite this article
Markolovic, S., Zhuang, Q., Wilkins, S.E. et al. The Jumonji-C oxygenase JMJD7 catalyzes (3S)-lysyl hydroxylation of TRAFAC GTPases. Nat Chem Biol 14, 688–695 (2018). https://doi.org/10.1038/s41589-018-0071-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0071-y
This article is cited by
-
Recent Advances and Therapeutic Implications of 2-Oxoglutarate-Dependent Dioxygenases in Ischemic Stroke
Molecular Neurobiology (2023)
-
Structure and function of cancer-related developmentally regulated GTP-binding protein 1 (DRG1) is conserved between sponges and humans
Scientific Reports (2022)
-
Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5
Scientific Reports (2022)
-
Conservation of the unusual dimeric JmjC fold of JMJD7 from Drosophila melanogaster to humans
Scientific Reports (2022)
-
Developmentally regulated GTPases: structure, function and roles in disease
Cellular and Molecular Life Sciences (2021)