Abstract
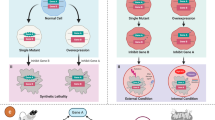
The concept of synthetic lethality has been widely applied to identify therapeutic targets in cancer, with varying degrees of success. The standard approach normally involves identifying genetic interactions between two genes, a driver and a target. In reality, however, most cancer synthetic lethal effects are likely complex and also polygenic, being influenced by the environment in addition to involving contributions from multiple genes. By acknowledging and delineating this complexity, we describe in this article how the success rate in cancer drug discovery and development could be improved.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ryan, C. J., Bajrami, I. & Lord, C. J. Synthetic lethality and cancer—penetrance as the major barrier. Trends Cancer 4, 671–683 (2018).
Fisher, R. A. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52, 399–433 (1919).
Kaelin, W. G. Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 (2005).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Behan, F. M. et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019).
Kryukov, G. V. et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 (2016).
Sulahian, R. et al. Synthetic lethal interaction of SHOC2 depletion with MEK inhibition in RAS-driven cancers. Cell Rep. 29, 118–134 (2019).
McDonald, E. R. III et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592 (2017).
Lord, C. J., Quinn, N. & Ryan, C. J. Integrative analysis of large-scale loss-of-function screens identifies robust cancer-associated genetic interactions. eLife 9, e58925 (2020).
Downward, J. RAS synthetic lethal screens revisited: still seeking the elusive prize? Clin. Cancer Res. 21, 1802–1809 (2015).
Kuzmin, E. et al. Systematic analysis of complex genetic interactions. Science 360, eaa01729 (2018).
Celaj, A. et al. Highly combinatorial genetic interaction analysis reveals a multi-drug transporter influence network. Cell Syst. 10, 25–38 (2020).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018).
Wicks, A. J., Krastev, D. B., Pettitt, S. J., Tutt, A. N. J. & Lord, C. J. Opinion: PARP inhibitors in cancer—what do we still need to know? Open Biol. 12, 220118 (2022).
Fahed, A. C. et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 11, 3635 (2020).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Brinch, S. A. et al. The tankyrase inhibitor OM-153 demonstrates antitumor efficacy and a therapeutic window in mouse models. Cancer Res. Commun. 2, 233–245 (2022).
Roberts, S. M. & Winston, F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465 (1997).
Ku, A. A. et al. Integration of multiple biological contexts reveals principles of synthetic lethality that affect reproducibility. Nat. Commun. 11, 2375 (2020).
Costanzo, M. et al. Environmental robustness of the global yeast genetic interaction network. Science 372, eabf8424 (2021).
Guenole, A. et al. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol. Cell 49, 346–358 (2013).
Herken, B. W., Wong, G. T., Norman, T. M. & Gilbert, L. A. Environmental challenge rewires functional connections among human genes. Preprint at bioRxiv https://doi.org/10.1101/2023.08.09.552346 (2023).
Rossiter, N. J. et al. CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. Cell Metab. 33, 1248–1263 (2021).
Han, K. et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 580, 136–141 (2020).
Martin, T. D. et al. The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 373, 1327–1335 (2021).
Olivieri, M. et al. A genetic map of the response to DNA damage in human cells. Cell 182, 481–496 (2020).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAFV600E inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Kopetz, S. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 381, 1632–1643 (2019).
El Tekle, G. et al. Co-occurrence and mutual exclusivity: what cross-cancer mutation patterns can tell us. Trends Cancer 7, 823–836 (2021).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017).
Smida, M. et al. MEK inhibitors block growth of lung tumours with mutations in ataxia-telangiectasia mutated. Nat. Commun. 7, 13701 (2016).
Chandler, R. L. et al. Coexistent ARID1A–PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 6, 6118 (2015).
Zatreanu, D. et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021).
Zhou, J. et al. A first-in-class polymerase θ inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2, 598–610 (2021).
Gatenby, R. A. & Brown, J. S. The evolution and ecology of resistance in cancer therapy. Cold Spring Harb. Perspect. Med. 10, a040972 (2020).
Dietlein, F. et al. A synergistic interaction between Chk1- and MK2 inhibitors in KRAS-mutant cancer. Cell 162, 146–159 (2015).
Ito, T. et al. Paralog knockout profiling identifies DUSP4 and DUSP6 as a digenic dependence in MAPK pathway-driven cancers. Nat. Genet. 53, 1664–1672 (2021).
Kelly, M. R. et al. Combined proteomic and genetic interaction mapping reveals new RAS effector pathways and susceptibilities. Cancer Discov. 10, 1950–1967 (2020).
Mencher, S. K. & Wang, L. G. Promiscuous drugs compared to selective drugs (promiscuity can be a virtue). BMC Clin. Pharmacol. 5, 3 (2005).
Pettitt, S. J. et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 10, 1475–1488 (2020).
Pettitt, S. J. et al. Genome-wide and high-density CRISPR–Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018).
Dunn, S. et al. AKT–mTORC1 reactivation is the dominant resistance driver for PI3Kβ/AKT inhibitors in PTEN-null breast cancer and can be overcome by combining with Mcl-1 inhibitors. Oncogene 41, 5046–5060 (2022).
Han, K. et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474 (2017).
Collins, S. R. et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 (2007).
Simpson, D., Ling, J., Jing, Y. & Adamson, B. Mapping the genetic interaction network of PARP inhibitor response. Preprint at bioRxiv https://doi.org/10.1101/2023.08.19.553986 (2023).
van Leeuwen, J., Boone, C. & Andrews, B. J. Mapping a diversity of genetic interactions in yeast. Curr. Opin. Syst. Biol. 6, 14–21 (2017).
Horlbeck, M. A. et al. Mapping the genetic landscape of human cells. Cell 174, 953–967 (2018).
Boettcher, M. et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat. Biotechnol. 36, 170–178 (2018).
Deans, R. M. et al. Parallel shRNA and CRISPR–Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 12, 361–366 (2016).
Ryan, C. J., Mehta, I., Kebabci, N. & Adams, D. J. Targeting synthetic lethal paralogs in cancer. Trends Cancer 9, 397–409 (2023).
Wang, J. et al. Computational methods, databases and tools for synthetic lethality prediction. Brief. Bioinform. 23, bbac106 (2022).
De Kegel, B., Quinn, N., Thompson, N. A., Adams, D. J. & Ryan, C. J. Comprehensive prediction of robust synthetic lethality between paralog pairs in cancer cell lines. Cell Syst. 12, 1144–1159 (2021).
Al-Anzi, B. F., Khajah, M. & Fakhraldeen, S. A. Predicting and explaining the impact of genetic disruptions and interactions on organismal viability. Bioinformatics 38, 4088–4099 (2022).
Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018).
Jaaks, P. et al. Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 603, 166–173 (2022).
Menden, M. P. et al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat. Commun. 10, 2674 (2019).
Posch, C. et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc. Natl Acad. Sci. USA 110, 4015–4020 (2013).
Kuenzi, B. M. et al. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell 38, 672–684 (2020).
Acknowledgements
We thank C. Astier for initial conversations regarding this article. We also thank the following for funding work in our respective laboratories: C.J.L. and S.J.P. thank Breast Cancer Now as part of program funding to the Breast Cancer Now Toby Robins Research Centre, Cancer Research UK as part of a program grant and the Basser Foundation. C.J.R. thanks the Science Foundation for funding under grant number 20/FFP-P/8641. Figures were generated using BioRender.
Author information
Authors and Affiliations
Contributions
The concept for this article was conceived by C.J.L. and C.J.R. The article and figures were written and compiled by C.J.L., C.J.R., L.P.S.D. and S.J.P.
Corresponding authors
Ethics declarations
Competing interests
C.J.L. makes the following disclosures: he receives and/or has received research funding from AstraZeneca, Merck and Artios; he received consultancy, SAB membership or honoraria payments from Syncona, Sun Pharma, Gerson Lehrman Group, Merck, Vertex, AstraZeneca, Tango, Third Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, NeoPhore and GlaxoSmithKline; he has stock in Tango, Ovibio, Hysplex and Tesselate. C.J.L. is also a named inventor on patents describing the use of DNA-repair inhibitors and stands to gain from their development and use as part of the ICR ‘Rewards to Inventors’ scheme and also reports benefits from this scheme associated with patents for PARPi paid into C.J.L.’s personal account and research accounts at the Institute of Cancer Research. C.J.R., L.P.S.D. and S.J.P. have no competing interests to declare.
Peer review information
Nature Genetics thanks William Sellers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ryan, C.J., Devakumar, L.P.S., Pettitt, S.J. et al. Complex synthetic lethality in cancer. Nat Genet 55, 2039–2048 (2023). https://doi.org/10.1038/s41588-023-01557-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-023-01557-x