Abstract
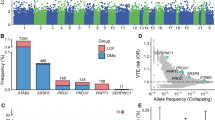
We report a genome-wide association study of venous thromboembolism (VTE) incorporating 81,190 cases and 1,419,671 controls sampled from six cohorts. We identify 93 risk loci, of which 62 are previously unreported. Many of the identified risk loci are at genes encoding proteins with functions converging on the coagulation cascade or platelet function. A VTE polygenic risk score (PRS) enabled effective identification of both high- and low-risk individuals. Individuals within the top 0.1% of PRS distribution had a VTE risk similar to homozygous or compound heterozygous carriers of the variants G20210A (c.*97 G > A) in F2 and p.R534Q in F5. We also document that F2 and F5 mutation carriers in the bottom 10% of the PRS distribution had a risk similar to that of the general population. We further show that PRS improved individual risk prediction beyond that of genetic and clinical risk factors. We investigated the extent to which venous and arterial thrombosis share clinical risk factors using Mendelian randomization, finding that some risk factors for arterial thrombosis were directionally concordant with VTE risk (for example, body mass index and smoking) whereas others were discordant (for example, systolic blood pressure and triglyceride levels).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
GWAS meta-analysis summary statistics and GWAS summary statistics from CHB-CVDC/DBDS, Intermountain healthcare and deCODE are available at https://www.decode.com/summarydata. Data from the UKB samples are available through UKB (https://www.ukbiobank.ac.uk/). FinnGen GWAS summary statistics are publicly accessible following registration (https://www.finngen.fi/en/access_results). Data from MVP are publicly available through dbGAP, accession code phs001672.v2.p1. The VTE PRS is available at the PGS Catalog (PGS ID accession: PGS003332). Individual-level data sharing is subject to restrictions imposed by patient consent and local ethics review boards. The GTEx v.8 eQTL data used in this study are available in the GTEx Portal (https://gtexportal.org/home/datasets). MSigDB gene sets are available online (https://www.gsea-msigdb.org/gsea/msigdb/) and through FUMA (https://fuma.ctglab.nl/).
Code availability
The following software and packages were used for data analysis: PLINK v.2.0 (https://www.cog-genomics.org/plink/2.0/), METAL v.2011-03-25 (http://csg.sph.umich.edu/abecasis/Metal/download/), MAGMA v.1.07 (https://ctg.cncr.nl/software/magma), EasyQC v.9.2 (https://www.uni-regensburg.de/medizin/epidemiologie-praeventivmedizin/genetische-epidemiologie/software/), FUMA v.1.3.8 (https://fuma.ctglab.nl/), LD score regression v.1.0.1 (https://github.com/bulik/ldsc), PoPS v.0.1 (https://github.com/FinucaneLab/pops/tree/add-license-1), PRS–CS v.2021-06-04 (https://github.com/getian107/PRScs/), REGENIE v.2.0.1 (https://rgcgithub.github.io/regenie/), TwoSampleMR v.0.5.6 (https://mrcieu.github.io/TwoSampleMR/), MiXeR v.1.3.0 (https://github.com/precimed/mixer), BOLT-REML v.2.4 (https://alkesgroup.broadinstitute.org/BOLT-LMM/BOLT-LMM_manual.html) and R v.4.1.2 (https://www.r-project.org/).
References
Cohen, A. T. et al. Venous thromboembolism (VTE) in Europe. Thromb. Haemost. 98, 756–764 (2007).
Beckman, M. G., Hooper, W. C., Critchley, S. E. & Ortel, T. L. Venous thromboembolism: a public health concern. Am. J. Prev. Med. 38, S495–S501 (2010).
Kahn, S. R. The post-thrombotic syndrome. Hematol. Am. Soc. Hematol. Educ. Program. 2016, 413–418 (2016).
Khan, F., Tritschler, T., Kahn, S. R. & Rodger, M. A. Venous thromboembolism. Lancet 398, 64–77 (2021).
Larsen, T. B. et al. Major genetic susceptibility for venous thromboembolism in men: a study of Danish twins. Epidemiology 14, 328–332 (2003).
Heit, J. A. et al. Familial segregation of venous thromboembolism. J. Thromb. Haemost. 2, 731–736 (2004).
Connors, J. M. Thrombophilia testing and venous thrombosis. N. Engl. J. Med. 377, 1177–1187 (2017).
Trégouët, D.-A. et al. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood 113, 5298–5303 (2009).
Buil, A. et al. C4BPB/C4BPA is a new susceptibility locus for venous thrombosis with unknown protein S-independent mechanism: results from genome-wide association and gene expression analyses followed by case-control studies. Blood 115, 4644–4650 (2010).
Heit, J. A. et al. Identification of unique venous thromboembolism-susceptibility variants in African-Americans. Thromb. Haemost. 117, 758–768 (2017).
Lindström, S. et al. Genomic and transcriptomic association studies identify 16 novel susceptibility loci for venous thromboembolism. Blood 134, 1645–1657 (2019).
Klarin, D. et al. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat. Genet. 51, 1574–1579 (2019).
Geerts, W. H. et al. Prevention of venous thromboembolism: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th edition). Chest 133, 381S–453S (2008).
Spyropoulos, A. C. et al. Predictive and associative models to identify hospitalized medical patients at risk for VTE. Chest 140, 706–714 (2011).
Barbar, S. et al. A risk assessment model for the identification of hospitalized medical patients at risk for venous thromboembolism: the Padua Prediction Score. J. Thromb. Haemost. 8, 2450–2457 (2010).
Caprini, J. A., Arcelus, J. I., Hasty, J. H., Tamhane, A. C. & Fabrega, F. Clinical assessment of venous thromboembolic risk in surgical patients. Semin. Thromb. Hemost. 17, 304–312 (1991). Suppl 3.
Bates, S. M. et al. American Society of Hematology 2018 guidelines for management of venous thromboembolism: venous thromboembolism in the context of pregnancy. Blood Adv. 2, 3317–3359 (2018).
Guyatt, G. H. et al. Introduction to the Ninth Edition: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest 141, 48S–52S (2012).
Martin, K. A. et al. Time trends in pulmonary embolism mortality rates in the United States, 1999 to 2018. J. Am. Heart Assoc. 9, e016784 (2020).
Vossen, C. Y. et al. Risk of a first venous thrombotic event in carriers of a familial thrombophilic defect. The European Prospective Cohort on Thrombophilia (EPCOT). J. Thromb. Haemost. 3, 459–464 (2005).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Mikaelsdottir, E. et al. Genetic variants associated with platelet count are predictive of human disease and physiological markers. Commun. Biol. 4, 1132 (2021).
Ferkingstad, E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021).
Sethi, M. K. et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J. Biol. Chem. 285, 1582–1586 (2010).
Liu, Y., Lee, J. W. & Ackerman, S. L. Mutations in the microtubule-associated protein 1A (Map1a) gene cause Purkinje cell degeneration. J. Neurosci. 35, 4587–4598 (2015).
Nikpay, M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Malik, R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 50, 524–537 (2018).
Klarin, D. et al. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat. Med. 25, 1274–1279 (2019).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Park, C. et al. Fasting glucose level and the risk of incident atherosclerotic cardiovascular diseases. Diabetes Care 36, 1988–1993 (2013).
Flint, A. C. et al. Effect of systolic and diastolic blood pressure on cardiovascular outcomes. N. Engl. J. Med. 381, 243–251 (2019).
Nordestgaard, B. G. & Varbo, A. Triglycerides and cardiovascular disease. Lancet 384, 626–635 (2014).
Klarin, D., Emdin, C. A., Natarajan, P., Conrad, M. F. & Kathiresan, S. Genetic analysis of venous thromboembolism in UK Biobank identifies the ZFPM2 locus and implicates obesity as a causal risk factor. Circ. Cardiovasc. Genet. 10, e001643 (2017).
Larsson, S. C. et al. Genetic predisposition to smoking in relation to 14 cardiovascular diseases. Eur. Heart J. 41, 3304–3310 (2020).
Allara, E. et al. Genetic determinants of lipids and cardiovascular disease outcomes. Circ. Genom. Precis. Med. 12, e002711 (2019).
Richardson, T. G. et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 17, e1003062 (2020).
de Haan, H. G. et al. Multiple SNP testing improves risk prediction of first venous thrombosis. Blood 120, 656–663 (2012).
Kujovich, J. L. Factor V Leiden thrombophilia. Genet. Med. 13, 1–16 (2011).
Weeks, E. M. et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. Preprint at https://www.medrxiv.org/content/10.1101/2020.09.08.20190561v1 (2020).
Jooss, N. J. et al. Role of platelet glycoprotein VI and tyrosine kinase Syk in thrombus formation on collagen-like surfaces. Int. J. Mol. Sci. 20, 2788 (2019).
Aslan, J. E., Tormoen, G. W., Loren, C. P., Pang, J. & McCarty, O. J. T. S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood 118, 3129–3136 (2011).
Louwette, S. et al. NPC1 defect results in abnormal platelet formation and function: studies in Niemann–Pick disease type C1 patients and zebrafish. Hum. Mol. Genet. 22, 61–73 (2013).
Pellegrini, V. D. Comparative effectiveness of pulmonary embolism prevention after hip and knee replacement: balancing safety and effectiveness. clinicaltrials.gov https://clinicaltrials.gov/ct2/show/NCT02810704 (2021).
Shivakumar, S. Extended venous thromboembolism prophylaxis comparing rivaroxaban and aspirin to aspirin alone following total hip and knee arthroplasty (EPCATIII). clinicaltrials.gov https://clinicaltrials.gov/ct2/show/NCT04075240 (2019).
Ortel, T. L. et al. American Society of Hematology 2020 guidelines for management of venous thromboembolism: treatment of deep vein thrombosis and pulmonary embolism. Blood Adv. 4, 4693–4738 (2020).
Mahmoodi, B. K. et al. Prevention of Renal and Vascular End-stage Disease (PREVEND) Study Group for the microalbuminuria and risk of venous thromboembolism. JAMA 301, 1790–1797 (2009).
Prandoni, P. et al. An association between atherosclerosis and venous thrombosis. N. Engl. J. Med. 348, 1435–1441 (2003).
Becattini, C. et al. A prospective study on cardiovascular events after acute pulmonary embolism. Eur. Heart J. 26, 77–83 (2005).
Holst, A. G., Jensen, G. & Prescott, E. Risk factors for venous thromboembolism: results from the Copenhagen City Heart Study. Circulation 121, 1896–1903 (2010).
Gregson, J. et al. Cardiovascular risk factors associated with venous thromboembolism. JAMA Cardiol. 4, 163–173 (2019).
Mahmoodi, B. K. et al. Association of traditional cardiovascular risk factors with venous thromboembolism. Circulation 135, 7–16 (2017).
Wang, H., Rosendaal, F. R., Cushman, M. & van Hylckama Vlieg, A. Association between cardiovascular risk factors and venous thromboembolism in the elderly. Res. Pract. Thromb. Haemost. 6, e12671 (2022).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Loh, P.-R. et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 47, 1385–1392 (2015).
Lee, S. H., Goddard, M. E., Wray, N. R. & Visscher, P. M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 36, 214–224 (2012).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Wallace, C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 16, e1008720 (2020).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Fahed, A. C. et al. Association of the interaction between familial hypercholesterolemia variants and adherence to a healthy lifestyle with risk of coronary artery disease. JAMA Netw. Open 5, e222687 (2022).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015).
Anderson, F. A. & Spencer, F. A. Risk factors for venous thromboembolism. Circulation 107, I9–I16 (2003).
Ageno, W., Becattini, C., Brighton, T., Selby, R. & Kamphuisen, P. W. Cardiovascular risk factors and venous thromboembolism. Circulation 117, 93–102 (2008).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011).
Pencina, M. J., D’Agostino, R. B., D’Agostino, R. B. & Vasan, R. S. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat. Med. 27, 157–172 (2008).
Pencina, M. J., D’Agostino, R. B. & Steyerberg, E. W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat. Med. 30, 11–21 (2011).
Hsu, C.-H. & Taylor, J. M. G. A robust weighted Kaplan-Meier approach for data with dependent censoring using linear combinations of prognostic covariates. Stat. Med. 29, 2215–2223 (2010).
Kundu, S., Aulchenko, Y. S., van Duijn, C. M. & Janssens, A. C. J. W. PredictABEL: an R package for the assessment of risk prediction models. Eur. J. Epidemiol. 26, 261–264 (2011).
Holland, D. et al. Beyond SNP heritability: polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet. 16, e1008612 (2020).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Acknowledgements
This research has been conducted using the UKB resource under application no. 43247. This work was supported by BRIDGE—Translational Excellence Programme (nos. NNF18SA0034956 and NNF20SA0064340), The John and Birthe Meyer Foundation, The Innovation Fund Denmark (PM Heart), NordForsk, Villadsen Family Foundation, The Arvid Nilsson Foundation, The Hallas-Møller Emerging Investigator Novo Nordisk (no. NNF17OC0031204) and Novo Nordisk Foundation (nos. NNF17OC0027594 and NNF14CC0001). All human research was approved within each contributing study by the relevant institutional review board (CHB-CVDC/DBDS: National Committee on Health Research Ethics; deCODE: National Bioethics Committee; Intermountain Healthcare: Intermountain Healthcare Institutional Review Board; UKB: Northwest Multicenter Research Ethics Committee; FinnGen: The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa; MVP: VA Central Institutional Review Board) and conducted according to the Declaration of Helsinki. All participants (except for CHB-CVDC) provided written informed consent. For CHB-CVDC, patients were informed about the opt-out possibility of having their biological specimens excluded from use in research in general. Since 2004, a national Register on Tissue Application (Vævsanvendelsesregistret) lists all individuals who have chosen to opt out and whose samples cannot be used for research purposes. Before initiating this study, individuals listed in the Register on Tissue Application were excluded. Additional information is provided in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
J.G., V.T., H.H., M.S.O. and H.B. conceived the study. J.G., V.T., E.F., G.A., G.H.H., S.G. and D.F.G. performed analyses in the respective cohorts. H.H., D.F.G., P.S., U.T., M.S.O. and H.B. supervised analyses in their respective cohorts. J.G., V.T., G.A., H.H., M.S.O. and H.B. contributed to writing the manuscript. J.G. and V.T. performed meta-analysis and created figures and tables. J.G and V.T. performed downstream analyses and drafted the manuscript. S.A.R., J.B.J., E.B.L., C.R.V., L.T., I.J., K.B., S.B., S.R.O., O.B.P., E.S., C.E., M.T.B., K.R.N., L.K., A.H.C., K.I., D.J., K.U.K., L.N., I.O., P.T.O., G.T. and K.S. interpreted the results and reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors who are affiliated with deCODE genetics/Amgen Inc. declare competing interests as employees. H.B. receives lecture fees from Bristol-Myers Squibb and Merck Sharp and Dohme. S.B. is a board member for Proscion A/S and Intomics A/S. All other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Bengt Zoller, Guillaume Pare and Norihiro Kato for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–3.
Supplementary Tables
Supplementary Tables 1–26.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghouse, J., Tragante, V., Ahlberg, G. et al. Genome-wide meta-analysis identifies 93 risk loci and enables risk prediction equivalent to monogenic forms of venous thromboembolism. Nat Genet 55, 399–409 (2023). https://doi.org/10.1038/s41588-022-01286-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01286-7
This article is cited by
-
FORGEdb: a tool for identifying candidate functional variants and uncovering target genes and mechanisms for complex diseases
Genome Biology (2024)
-
Genetic associations of protein-coding variants in venous thromboembolism
Nature Communications (2024)
-
Genetic correlations, shared risk genes and immunity landscapes between COVID-19 and venous thromboembolism: evidence from GWAS and bulk transcriptome data
Inflammation Research (2024)
-
Evaluating significance of European-associated index SNPs in the East Asian population for 31 complex phenotypes
BMC Genomics (2023)
-
Identifying individuals at extreme risk of venous thromboembolism using polygenic risk scores
Nature Genetics (2023)