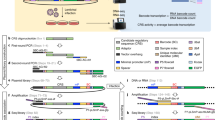
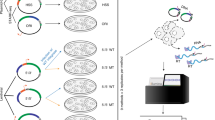
Abstract
Massively parallel reporter gene assays are key tools in regulatory genomics but cannot be used to identify cell-type-specific regulatory elements without performing assays serially across different cell types. To address this problem, we developed a single-cell massively parallel reporter assay (scMPRA) to measure the activity of libraries of cis-regulatory sequences (CRSs) across multiple cell types simultaneously. We assayed a library of core promoters in a mixture of HEK293 and K562 cells and showed that scMPRA is a reproducible, highly parallel, single-cell reporter gene assay that detects cell-type-specific cis-regulatory activity. We then measured a library of promoter variants across multiple cell types in live mouse retinas and showed that subtle genetic variants can produce cell-type-specific effects on cis-regulatory activity. We anticipate that scMPRA will be widely applicable for studying the role of CRSs across diverse cell types.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Next-generation sequencing data that support the findings of the study are available in the Gene Expression Omnibus using accession code GSE188639.
Code availability
The code that supports the findings of this study is available in Zenodo52.
References
Schaub, M. A., Boyle, A. P., Kundaje, A., Batzoglou, S. & Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Hindorff, L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367 (2009).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Vattikuti, S., Guo, J. & Chow, C. C. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 8, e1002637 (2012).
Shi, H., Kichaev, G. & Pasaniuc, B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 99, 139–153 (2016).
Aygün, N. et al. Brain-trait-associated variants impact cell-type-specific gene regulation during neurogenesis. Am. J. Hum. Genet. 108, 1647–1668 (2021).
Nott, A. et al. Brain cell type-specific enhancer–promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019).
Spielmann, M. & Mundlos, S. Looking beyond the genes: the role of non-coding variants in human disease. Hum. Mol. Genet. 25, R157–R165 (2016).
Zhang, F. & Lupski, J. R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 24, R102–R110 (2015).
Ong, C.-T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011).
Arnold, C. D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013).
Kwasnieski, J. C., Mogno, I., Myers, C. A., Corbo, J. C. & Cohen, B. A. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl Acad. Sci. USA 109, 19498–19503 (2012).
Ireland, W.T. et al. Deciphering the regulatory genome of Escherichia coli, one hundred promoters at a time. eLife 9, e55308 (2020).
Patwardhan, R. P. et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30, 265–270 (2012).
Sharon, E. et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530 (2012).
Kinney, J. B., Murugan, A., Callan, C. G. Jr & Cox, E. C. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc. Natl Acad. Sci. USA 107, 9158–9163 (2010).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 (2012).
White, M. A. et al. A simple grammar defines activating and repressing cis-regulatory elements in photoreceptors. Cell Rep. 17, 1247–1254 (2016).
Kwasnieski, J. C., Fiore, C., Chaudhari, H. G. & Cohen, B. A. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 24, 1595–1602 (2014).
Chaudhari, H. G. & Cohen, B. A. Local sequence features that influence AP-1 cis-regulatory activity. Genome Res. 28, 171–181 (2018).
Hughes, A. E. O., Myers, C. A. & Corbo, J. C. A massively parallel reporter assay reveals context-dependent activity of homeodomain binding sites in vivo. Genome Res. 28, 1520–1531 (2018).
Tewhey, R. et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016).
Hong, C. K. Y. & Cohen, B. A. Genomic environments scale the activities of diverse core promoters. Genome Res. 32, 85–96 (2022).
Haberle, V. et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570, 122–126 (2019).
Zabidi, M. A. et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017).
Moudgil, A. et al. Self-reporting transposons enable simultaneous readout of gene expression and transcription factor binding in single cells. Cell 182, 992–1008 (2020).
Litzenburger, U. M. et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol. 18, 15 (2017).
Min, M. & Spencer, S. L. Spontaneously slow-cycling subpopulations of human cells originate from activation of stress-response pathways. PLoS Biol. 17, e3000178 (2019).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Ishikawa, F. et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 25, 1315–1321 (2007).
Friedman, R. Z. et al. Information content differentiates enhancers from silencers in mouse photoreceptors. eLife 10, e67403 (2021).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Murphy, D. P., Hughes, A. E., Lawrence, K. A., Myers, C. A. & Corbo, J. C. Cis-regulatory basis of sister cell type divergence in the vertebrate retina. eLife 8, e48216 (2019).
Reese, B. E. Development of the retina and optic pathway. Vis. Res. 51, 613–632 (2011).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Bryant, D. H. et al. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 39, 691–696 (2021).
Chan, Y. K. et al. Engineering adeno-associated viral vectors to evade innate immune and inflammatory responses. Sci. Transl. Med. 13, eabd3438 (2021).
Byrne, L. C. et al. In vivo-directed evolution of adeno-associated virus in the primate retina. JCI Insight 5, e135112 (2020).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Shen, S. Q. et al. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 26, 238–255 (2016).
Cohen, R. N., van der Aa, M. A. E. M., Macaraeg, N., Lee, A. P. & Szoka, F. C. Jr. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release 135, 166–174 (2009).
Hsiau, T. H.-C. et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One 2, e643 (2007).
Montana, C. L., Myers, C. A. & Corbo, J. C. Quantifying the activity of cis-regulatory elements in the mouse retina by explant electroporation. J. Vis. Exp.(52), 2821 (2011).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Bailey, T. L. & Gribskov, M. Combining evidence using P values: application to sequence homology searches. Bioinformatics 14, 48–54 (1998).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Waskom, M. Seaborn: statistical data visualization. J. Open Source Softw. 6, 3021 (2021).
Zhao, S. et al. A single-cell massively parallel reporter assay detects cell type specific cis-regulatory activity. https://doi.org/10.5281/zenodo.7338678 (2022).
Acknowledgements
We thank the members of the Cohen laboratory for their critical feedback on the manuscript. We thank J. Hoisington-Lopez and M. Crosby for assistance with high-throughput sequencing. This work is supported by grants to B.A.C. from the National Institutes of Health (R01 GM140711 and R01 GM092910) and to J.C.C. from the National Institutes of Health (R01 EY030075). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.Z. and B.A.C. conceived and designed the project. All experiments and analyses were performed by S.Z. with technical contributions from C.K.Y. H. and D.M.G., except for the electroporation and culturing of mouse retinas, which was performed by C.A.M. J.C.C and M.A.W provided critical input into the design of the Gnb3 promoter variant library. S.Z., C.K.Y.H. and B.A.C. wrote the manuscript with input and feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
S.Z. and B.A.C. are inventors on a pending patent filed by Washington University in St. Louis which may encompass the methods, reagents and data disclosed in this manuscript. B.A.C. is on the scientific advisory board of Patch Biosciences. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Bas van Steensel, Rickard Sandberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 scMPRA measures cell-type specific CRS activity.
(a) UMAP of the single-cell transcriptome from the mixed-cell experiment. 105 out of 3417 cells (3%) are labeled by both K562 and HEK293 cell genes. (b) UMAP of the mixed-cell experiment with cells marked by other representative markers for K562 and HEK293 cell expression. (c, d) Histogram of the number of plasmids (unique cBC-rBC pairs) transfected into K562 cells and HEK293 cells. (e, f) Histogram of the mean number of rBC per cBC (CRS) per cell for K562 cells and HEK293 cells. (g, h) Correlation of bulk MPRA versus scMPRA where only the scMPRA data has been UMI normalized (i, j) Scatterplot of scMPRA reproducibility for housekeeping and developmental promoters in K562 cells and HEK293 cells.
Extended Data Fig. 2 scMPRA measures CRS activity in K562 cell substates.
(a) Reproducibility for mean expression of core promoters in K562 cells. (b) Correlation of bulk and scMPRA (non-UMI corrected) in K562 cells (c) Different dynamics of expression. For UBA52, the promoter is most highly expressed in S phase, whereas for CSF1, the promoter is most highly expressed in G1 phase. For CXCL10, the promoter is expressed evenly through cell cycle (Stars indicate significance from two-sided Wilcoxon rank sum test, *: p < 0.05) (d) Cells no longer cluster together based on cell cycle genes after the effects of the cell cycle are removed.
Extended Data Fig. 3 Robust measurements of Gnb3 promoter library in ex vivo retina.
(a) Expression of marker genes by scRNA-seq used to identify cell types in the retina. (b) Percentage of the total cells recovered represented by each retinal cell type. (c) Plot showing the relationship between the mean activity of a Gnb3 promoter variant in a given cell type (x-axis) and the proportion of cells in which that promoter variant is silent (y-axis). Individual cells in which a given Gnb3 variant is silent are identified as cells with U6-expressed cBC, but no Gnb3-expressed cBC. (d) The correlation between biological replicates (n = 2) is plotted as a function of the number of cells used in the analysis. The bounds of the box represent the upper and lower quartiles respectively, and the center line represents the median. The whiskers extend to the maxima/minima except for points determined to be outliers using a method that is a function of the interquartile range.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Table 1—Mixed-cell experiment expression. Supplementary Table 2—Differential expression of the core promoter library between K562 and HEK293 cells. Supplementary Table 3—K562 cell cycle expression. Supplementary Table 4—K562 cell substate expression. Supplementary Table 5—Gnb3 promoter variant library. Supplementary Table 6—Gnb3 library expression in retina. Supplementary Table 7—Oligos used in this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, S., Hong, C.K.Y., Myers, C.A. et al. A single-cell massively parallel reporter assay detects cell-type-specific gene regulation. Nat Genet 55, 346–354 (2023). https://doi.org/10.1038/s41588-022-01278-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01278-7
This article is cited by
-
Characterization of enhancer activity in early human neurodevelopment using Massively Parallel Reporter Assay (MPRA) and forebrain organoids
Scientific Reports (2024)
-
Hold out the genome: a roadmap to solving the cis-regulatory code
Nature (2024)
-
Decoding enhancer complexity with machine learning and high-throughput discovery
Genome Biology (2023)