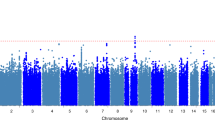
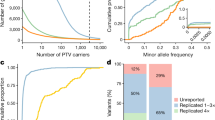
Abstract
Osteoarthritis is a common progressive joint disease. As no effective medical interventions are available, osteoarthritis often progresses to the end stage, in which only surgical options such as total joint replacement are available. A more thorough understanding of genetic influences of osteoarthritis is essential to develop targeted personalized approaches to treatment, ideally long before the end stage is reached. To date, there have been no large multiancestry genetic studies of osteoarthritis. Here, we leveraged the unique resources of 484,374 participants in the Million Veteran Program and UK Biobank to address this gap. Analyses included participants of European, African, Asian and Hispanic descent. We discovered osteoarthritis-associated genetic variation at 10 loci and replicated findings from previous osteoarthritis studies. We also present evidence that some osteoarthritis-associated regions are robust to population ancestry. Drug repurposing analyses revealed enrichment of targets of several medication classes and provide potential insight into the etiology of beneficial effects of antiepileptics on osteoarthritis pain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The MVP GWAS summary statistics generated and/or analyzed during the current study will be made available via dbGAP; the dbGaP accession assigned to the MVP is phs001672.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v7.p1). Summary statistics from the UKB results are available in the GWAS catalog (https://www.ebi.ac.uk/gwas/home) under the following accessions: GCST90134279, GCST90134280, GCST90134281, GCST90134282, GCST90134283, GCST90134284, GCST90134285, GCST90134286, GCST90134287, GCST90134288, GCST90134289, GCST90134290.
References
Yoon, J., Scott, J. Y., Phibbs, C. S. & Wagner, T. H. Recent trends in Veterans Affairs chronic condition spending. Popul. Health Manag. 14, 293–298 (2011).
Musumeci, G. et al. Osteoarthritis in the XXIst century: risk factors and behaviours that influence disease onset and progression. Int. J. Mol. Sci. 16, 6093–6112 (2015).
Singh, J. A. & Lewallen, D. Age, gender, obesity, and depression are associated with patient-related pain and function outcome after revision total hip arthroplasty. Clin. Rheumatol. 28, 1419–1430 (2009).
Warner, S. & Valdes, A. The genetics of osteoarthritis: a review. J. Funct. Morphol. Kinesiol. 1, 140–153 (2016).
Castano Betancourt, M. C. et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl Acad. Sci. USA 109, 8218–8223 (2012).
Evangelou, E. et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann. Rheum. Dis. 73, 2130–2136 (2014).
Evans, D. S. et al. Genome-wide association and functional studies identify a role for IGFBP3 in hip osteoarthritis. Ann. Rheum. Dis. 74, 1861–1867 (2015).
Styrkarsdottir, U. et al. GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat. Commun. 10, 2054 (2019).
Styrkarsdottir, U. et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat. Genet. 46, 498–502 (2014).
Valdes, A. M. et al. Genome-wide association scan identifies a prostaglandin-endoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am. J. Hum. Genet. 82, 1231–1240 (2008).
Zeggini, E. et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380, 815–823 (2012).
Reynard, L. N. & Barter, M. J. Osteoarthritis year in review 2019: genetics, genomics and epigenetics. Osteoarthritis Cartilage 28, 275–284 (2020).
Boer, C. G. et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 184, 4784–4818.e17 (2021).
Tachmazidou, I. et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank. Nat. Genet. 51, 230–236 (2019).
Styrkarsdottir, U. et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat. Genet. 50, 1681–1687 (2018).
Yau, M. S. et al. Genome-wide association study of radiographic knee osteoarthritis in North American Caucasians. Arthritis Rheumatol. 69, 343–351 (2017).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Montagne, K. et al. High hydrostatic pressure induces pro-osteoarthritic changes in cartilage precursor cells: a transcriptome analysis. PLoS ONE 12, e0183226 (2017).
Khan, A. T. et al. Recommendations on the use and reporting of race, ethnicity, and ancestry in genetic research: experiences from the NHLBI TOPMed program. Cell Genomics 2, 100155 (2022).
Chu, M. L. & Tsuda, T. Fibulins in development and heritable disease. Birth Defects Res. C Embryo Today 72, 25–36 (2004).
Atef Nassif, M. Fibulin-3 serum and urine levels in the diagnosis and severity assessment of primary knee osteoarthritis. Reumatologia 57, 271–276 (2019).
Runhaar, J., Sanchez, C., Taralla, S., Henrotin, Y. & Bierma-Zeinstra, S. M. Fibulin-3 fragments are prognostic biomarkers of osteoarthritis incidence in overweight and obese women. Osteoarthritis Cartilage 24, 672–678 (2016).
Saberi Hosnijeh, F., Bierma-Zeinstra, S. M. & Bay-Jensen, A. C. Osteoarthritis year in review 2018: biomarkers (biochemical markers). Osteoarthritis Cartilage 27, 412–423 (2019).
Bakilan, F. et al. Effects of native type II collagen treatment on knee osteoarthritis: a randomized controlled trial. Eurasian J. Med. 48, 95–101 (2016).
Bradley, E. W. et al. Phlpp1 facilitates post-traumatic osteoarthritis and is induced by inflammation and promoter demethylation in human osteoarthritis. Osteoarthritis Cartilage 24, 1021–1028 (2016).
Bennett, D. L. H. & Woods, C. G. Painful and painless channelopathies. Lancet Neurol. 13, 587–599 (2014).
Taylor, S. E., Li, Y. H., Wong, W. H. & Bhutani, N. Genome-wide mapping of DNA hydroxymethylation in osteoarthritic chondrocytes. Arthritis Rheumatol. 67, 2129–2140 (2015).
Arshad, F. & Bishop, N. Osteogenesis imperfecta in children. Bone 148, 115914 (2021).
Garbazza, C. & Benedetti, F. Genetic factors affecting seasonality, mood, and the circadian clock. Front. Endocrinol. 9, 481 (2018).
Berenbaum, F. & Meng, Q. J. The brain-joint axis in osteoarthritis: nerves, circadian clocks and beyond. Nat. Rev. Rheumatol. 12, 508–516 (2016).
Eisenhauer, J. G. Meta‐analysis and mega‐analysis: a simple introduction. Teach. Stat. 43, 21–27 (2020).
Boedhoe, P. S. W. et al. An empirical comparison of meta- and mega-analysis with data from the ENIGMA Obsessive-Compulsive Disorder Working Group. Front. Neuroinform. 12, 102 (2018).
Zhao, X. et al. Whole genome sequence analysis of pulmonary function and COPD in 19,996 multi-ethnic participants. Nat. Commun. 11, 5182 (2020).
Little, A. et al. Whole genome sequence analysis of platelet traits in the NHLBI Trans-Omics for Precision Medicine (TOPMed) initiative. Hum. Mol. Genet. 31, 347–361 (2022).
Sarnowski, C. et al. Impact of rare and common genetic variants on diabetes diagnosis by hemoglobin A1c in multi-ancestry cohorts: the Trans-Omics for Precision Medicine Program. Am. J. Hum. Genet. 105, 706–718 (2019).
Taub, M. A. et al. Genetic determinants of telomere length from 109,122 ancestrally diverse whole-genome sequences in TOPMed. Cell Genomics 2, 100084 (2022).
Davis, J. P. et al. Common, low-frequency, and rare genetic variants associated with lipoprotein subclasses and triglyceride measures in Finnish men from the METSIM study. PLoS Genet. 13, e1007079 (2017).
Farnaghi, S., Crawford, R., Xiao, Y. & Prasadam, I. Cholesterol metabolism in pathogenesis of osteoarthritis disease. Int. J. Rheum. Dis. 20, 131–140 (2017).
Rask-Andersen, M., Karlsson, T., Ek, W. E. & Johansson, A. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 10, 339 (2019).
Bai, W. Y. et al. Integrative analysis of genomic and epigenomic data reveal underlying superenhancer-mediated microRNA regulatory network for human bone mineral density. Hum. Mol. Genet. 30, 2177–2189 (2021).
Weissbrod, O. et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 54, 450–458 (2022).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021).
Tachmazidou, I. et al. Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am. J. Hum. Genet. 100, 865–884 (2017).
Kettunen, J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 7, 11122 (2016).
Hartley, A. et al. Using multivariable Mendelian randomization to estimate the causal effect of bone mineral density on osteoarthritis risk, independently of body mass index. Int. J. Epidemiol. 51, 1254–1267 (2022).
Funck-Brentano, T., Nethander, M., Moverare-Skrtic, S., Richette, P. & Ohlsson, C. Causal factors for knee, hip, and hand osteoarthritis: a Mendelian randomization study in the UK Biobank. Arthritis Rheumatol. 71, 1634–1641 (2019).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Zhu, Z. et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 145, 537–549 (2020).
Ripatti, P. et al. Polygenic hyperlipidemias and coronary artery disease risk. Circ. Genom. Precis. Med. 13, e002725 (2020).
Liu, C. et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. 48, 1162–1170 (2016).
Han, X. et al. Using Mendelian randomization to evaluate the causal relationship between serum C-reactive protein levels and age-related macular degeneration. Eur. J. Epidemiol. 35, 139–146 (2020).
Sinnott-Armstrong, N. et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021).
Gratten, J. & Visscher, P. M. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med. 8, 78 (2016).
Solovieff, N., Cotsapas, C., Lee, P. H., Purcell, S. M. & Smoller, J. W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013).
Paaby, A. B. & Rockman, M. V. The many faces of pleiotropy. Trends Genet. 29, 66–73 (2013).
Stearns, F. W. One hundred years of pleiotropy: a retrospective. Genetics 186, 767–773 (2010).
van den Driest, J. J. et al. Antidepressant and anticonvulsant prescription rates in patients with osteoarthritis: a population-based cohort study. Rheumatology 60, 2206–2216 (2021).
Piersma, B., Hayward, M. K. & Weaver, V. M. Fibrosis and cancer: a strained relationship. Biochim. Biophys. Acta Rev. Cancer 1873, 188356 (2020).
Yazdani, S., Bansal, R. & Prakash, J. Drug targeting to myofibroblasts: implications for fibrosis and cancer. Adv. Drug Deliv. Rev. 121, 101–116 (2017).
Bay-Jensen, A. C., Thudium, C. S. & Mobasheri, A. Development and use of biochemical markers in osteoarthritis: current update. Curr. Opin. Rheumatol. 30, 121–128 (2018).
Yin, H. et al. Metabolomic analysis of biochemical changes in urine of osteoarthritis rat and interventional effects of Bushen-Huoxue herb couple. Chin. Herbal Med. 9, 369–375 (2017).
Valdes, A. M. et al. Association of beta-blocker use with less prevalent joint pain and lower opioid requirement in people with osteoarthritis. Arthritis Care Res. 69, 1076–1081 (2017).
Singhmar, P. et al. Critical role for Epac1 in inflammatory pain controlled by GRK2-mediated phosphorylation of Epac1. Proc. Natl Acad. Sci. USA 113, 3036–3041 (2016).
Evron, T., Daigle, T. L. & Caron, M. G. GRK2: multiple roles beyond G protein-coupled receptor desensitization. Trends Pharmacol. Sci. 33, 154–164 (2012).
Styrkarsdottir, U. et al. Whole-genome sequencing identifies rare genotypes in COMP and CHADL associated with high risk of hip osteoarthritis. Nat. Genet. 49, 801–805 (2017).
Fang, H. et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am. J. Hum. Genet. 105, 763–772 (2019).
Bennell, K. L., Hunter, D. J. & Hinman, R. S. Management of osteoarthritis of the knee. BMJ 345, e4934 (2012).
Whittaker, J. L., Runhaar, J., Bierma-Zeinstra, S. & Roos, E. M. A lifespan approach to osteoarthritis prevention. Osteoarthritis Cartilage 29, 1638–1653 (2021).
Murphy, L. B. et al. Arthritis among veterans – United States, 2011–2013. MMWR Morb. Mortal. Wkly Rep. 63, 999–1003 (2014).
Stanishewski, M. & Zimmermann, B. Osteoarthritis treatment in the veteran population. Fed. Pract. 32, 21S–25S (2015).
Hinojosa, R. & Hinojosa, M. S. Activity-limiting musculoskeletal conditions in US veterans compared to non-veterans: results from the 2013 National Health Interview Survey. PLoS ONE 11, e0167143 (2016).
Toivanen, A. T. et al. Obesity, physically demanding work and traumatic knee injury are major risk factors for knee osteoarthritis—a population-based study with a follow-up of 22 years. Rheumatology 49, 308–314 (2010).
Lin, D. Y. A simple and accurate method to determine genomewide significance for association tests in sequencing studies. Genet. Epidemiol. 43, 365–372 (2019).
Sobota, R. S. et al. Addressing population-specific multiple testing burdens in genetic association studies. Ann. Hum. Genet. 79, 136–147 (2015).
Asif, H. et al. GWAS significance thresholds for deep phenotyping studies can depend upon minor allele frequencies and sample size. Mol. Psychiatry 26, 2048–2055 (2021).
Chen, Z., Boehnke, M., Wen, X. & Mukherjee, B. Revisiting the genome-wide significance threshold for common variant GWAS. G3 11, jkaa056 (2021).
Zengini, E. et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat. Genet. 50, 549–558 (2018).
Hunter-Zinck, H. et al. Genotyping array design and data quality control in the Million Veteran Program. Am. J. Hum. Genet. 106, 535–548 (2020).
Morris, A. P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 35, 809–822 (2011).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Kichaev, G. & Pasaniuc, B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97, 260–271 (2015).
Kichaev, G. et al. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics 33, 248–255 (2017).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Kichaev, G. PAINTOR running software and suggested pipeline: https://github.com/gkichaev/PAINTOR_V3.0/wiki/3.-Running-Software-and-Suggested-Pipeline (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Speed, D., Holmes, J. & Balding, D. J. Evaluating and improving heritability models using summary statistics. Nat. Genet. 52, 458–462 (2020).
Sakaue, S. & Okada, Y. GREP: Genome for REPositioning drugs. Bioinformatics 35, 3821–3823 (2019).
Acknowledgements
We thank R. Tindal, a medical student in the McDonald laboratory at UAB, for assistance with manuscript editing, and C. Cole, a summer student in the McDonald laboratory at UAB, for assistance with formatting data. We thank the VA MVP, including members of the MVP Executive Committee, MVP Recruitment/Enrollment, MVP Science and current MVP Local Site Investigators, for providing the datasets used for the MVP analyses. We also thank the UKB for providing the datasets used for the UKB analyses. Please see the Supplementary Note for complete acknowledgement information. This research was based on data from the MVP, Office of Research and Development, Veterans Health Administration, and was supported by award no. I01RX002745 (M.M.B. and J.A.S.). This publication does not represent the views of the Department of Veteran Affairs or the United States Government.
Author information
Authors and Affiliations
Contributions
All authors contributed substantially to this manuscript. M.-L.N.M., J.S.R., R.D., V.J., C.J.B., H.K.T., M.M.B. and J.A.S. conceived and designed the study. P.L.K., V.S., A.N., A.P.R., S.A.P. and A.C.W. executed and summarized analyses. J.C. summarized analyses. H.K.T. and S.P. advised on statistical models and computational implementation. M.-L.N.M. wrote and revised the manuscript, which all authors reviewed and edited.
Corresponding author
Ethics declarations
Competing interests
All authors declare competing financial interests in the form of employment at UAB and/or US Veterans Health Administration. M.-L.N.M., J.C.R., R.D., C.J.B., S.P., H.K.T., J.A.S. and M.M.B. have received research support from the National Institutes of Health. J.A.S. has received consultant fees from Crealta/Horizon, Medisys, Fidia, UBM LLC, Trio health, Medscape, WebMD, PK Med, Two labs Inc., Adept Field Solutions, Clinical Care options, Clearview healthcare partners, Putnam associates, Focus forward, Navigant consulting, Spherix, Practice Point communications, the National Institutes of Health and the American College of Rheumatology. J.A.S. has received institutional research support from Zimmer Biomet Holdings. J.A.S. received food and beverage payments from Intuitive Surgical Inc./Philips Electronics North America. J.A.S. owns stock options in TPT Global Tech, Vaxart Pharmaceuticals, Atyu Biopharma, Adaptimmune Therapeutics, GeoVax Labs, Pieris Pharmaceuticals, Enzolytics Inc., Seres Therapeutics, Tonix Pharmaceuticals Holding Corp. and Charlotte’s Web Holdings, Inc. J.A.S. previously owned stock options in Amarin, Viking and Moderna pharmaceuticals. J.A.S. is on the speakers’ bureau of Simply Speaking. J.A.S. is a member of the executive of Outcomes Measures in Rheumatology, an organization that develops outcome measures in rheumatology and receives arms-length funding from 12 companies. J.A.S. serves on the FDA Arthritis Advisory Committee. J.A.S. is the chair of the Veterans Affairs Rheumatology Field Advisory Committee. J.A.S. is the editor and the Director of the UAB Cochrane Musculoskeletal Group Satellite Center on Network Meta-analysis. J.A.S. previously served on the following committees: member of the American College of Rheumatology (ACR) Annual Meeting Planning Committee and Quality of Care Committees, Chair of the ACR Meet-the-Professor, Workshop and Study Group Subcommittee, and cochair of the ACR Criteria and Response Criteria subcommittee.
Peer review
Peer review information
Nature Genetics thanks Unnur Styrkarsdottir and Fernando Rivadeneira for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–12.
Supplementary Table 1
Supplementary Tables 1–21.
Rights and permissions
About this article
Cite this article
McDonald, ML.N., Lakshman Kumar, P., Srinivasasainagendra, V. et al. Novel genetic loci associated with osteoarthritis in multi-ancestry analyses in the Million Veteran Program and UK Biobank. Nat Genet 54, 1816–1826 (2022). https://doi.org/10.1038/s41588-022-01221-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01221-w