Abstract
Esophageal squamous cell carcinomas (ESCCs) harbor recurrent chromosome 3q amplifications that target the transcription factor SOX2. Beyond its role as an oncogene in ESCC, SOX2 acts in development of the squamous esophagus and maintenance of adult esophageal precursor cells. To compare Sox2 activity in normal and malignant tissue, we developed engineered murine esophageal organoids spanning normal esophagus to Sox2-induced squamous cell carcinoma and mapped Sox2 binding and the epigenetic and transcriptional landscape with evolution from normal to cancer. While oncogenic Sox2 largely maintains actions observed in normal tissue, Sox2 overexpression with p53 and p16 inactivation promotes chromatin remodeling and evolution of the Sox2 cistrome. With Klf5, oncogenic Sox2 acquires new binding sites and enhances activity of oncogenes such as Stat3. Moreover, oncogenic Sox2 activates endogenous retroviruses, inducing expression of double-stranded RNA and dependence on the RNA editing enzyme ADAR1. These data reveal SOX2 functions in ESCC, defining targetable vulnerabilities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author. ChIP–seq, ATAC–seq, exome and RNA-seq data generated in this study were deposited to Gene Expression Omnibus (GEO) under the series GSE167367. Source data are provided with this paper.
Code availability
All software and bioinformatic tools used in the present study are publicly available.
References
Bass, A. J. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242 (2009).
Campbell, J. D. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689 e3 (2018).
Campbell, J. D. et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 23, 194–212 e6 (2018).
Dotto, G. P. & Rustgi, A. K. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell 29, 622–637 (2016).
Cancer Genome Atlas Research, N. et al. Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 (2017).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Wernig, M. et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 (2007).
Avilion, A. A. et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 (2003).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Masui, S. et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 (2007).
Que, J. et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531 (2007).
Yuan, P. et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS ONE 5, e9112 (2010).
Watanabe, H. et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J. Clin. Invest. 124, 1636–1645 (2014).
Garraway, L. A. & Sellers, W. R. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 6, 593–602 (2006).
Sulahian, R. et al. An integrative analysis reveals functional targets of GATA6 transcriptional regulation in gastric cancer. Oncogene 33, 5637–5648 (2014).
Salari, K. et al. CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proc. Natl Acad. Sci. USA 109, E3196–E3205 (2012).
Garraway, L. A. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122 (2005).
Adler, E. K. et al. The PAX8 cistrome in epithelial ovarian cancer. Oncotarget 8, 108316–108332 (2017).
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014).
Siegle, J. M. et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat. Commun. 5, 4511 (2014).
Justilien, V. et al. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 25, 139–151 (2014).
Ferone, G. et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 30, 519–532 (2016).
Mukhopadhyay, A. et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep. 8, 40–49 (2014).
Lazarus, K. A. et al. BCL11A interacts with SOX2 to control the expression of epigenetic regulators in lung squamous carcinoma. Nat. Commun. 9, 3327 (2018).
Daniely, Y. et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am. J. Physiol. Cell Physiol. 287, C171–C181 (2004).
Jiang, Y. et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 9, 3619 (2018).
Jiang, Y. Y. et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology 159, 1311–1327.e19 (2020).
Wang, S. et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8, 2502–2515 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Zhang, X. et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 48, 176–182 (2016).
Zhang, X. et al. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov. 8, 108–125 (2018).
Rogerson, C. et al. Repurposing of KLF5 activates a cell cycle signature during the progression from a precursor state to oesophageal adenocarcinoma. eLife 9, e57189 (2020).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 e16 (2017).
Gannon, H. S. et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat. Commun. 9, 5450 (2018).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019).
Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019).
Canadas, I. et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 24, 1143–1150 (2018).
Dodonova, S. O., Zhu, F., Dienemann, C., Taipale, J. & Cramer, P. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580, 669–672 (2020).
Liu, X. et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 14, 1169–1180 (2016).
Lee, H., Jeong, A. J. & Ye, S. K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 52, 415–423 (2019).
Karakasheva, T. A. et al. IL-6 mediates cross-talk between tumor cells and activated fibroblasts in the tumor microenvironment. Cancer Res. 78, 4957–4970 (2018).
Tarapore, R. S., Yang, Y. & Katz, J. P. Restoring KLF5 in esophageal squamous cell cancer cells activates the JNK pathway leading to apoptosis and reduced cell survival. Neoplasia 15, 472–480 (2013).
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650 e14 (2017).
Karin, M. & Clevers, H. Reparative inflammation takes charge of tissue regeneration. Nature 529, 307–315 (2016).
McConnell, B. B. et al. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology 140, 540–549 e2 (2011).
Mu, X., Ahmad, S. & Hur, S. Endogenous retroelements and the host innate immune sensors. Adv. Immunol. 132, 47–69 (2016).
Zeng, M. et al. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science 346, 1486–1492 (2014).
Baudino, L., Yoshinobu, K., Morito, N., Santiago-Raber, M. L. & Izui, S. Role of endogenous retroviruses in murine SLE. Autoimmun. Rev. 10, 27–34 (2010).
Ohnuki, M. et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl Acad. Sci. USA 111, 12426–12431 (2014).
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 e19 (2018).
Liddicoat, B. J. et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015).
Qin, Y. R. et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 74, 840–851 (2014).
Mehdipour, P. et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Zhang, X., Cowper-Sal lari, R., Bailey, S. D., Moore, J. H. & Lupien, M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 22, 1437–1446 (2012).
Qin, Q. et al. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC Bioinformatics 17, 404 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Liu, T. Use Model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein–DNA interactions in embryonic stem cells. Methods Mol. Biol. 1150, 81–95 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Cornwell, M. et al. VIPER: Visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics 19, 135 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kopylova, E., Noe, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Jin, Y., Tam, O. H., Paniagua, E. & Hammell, M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599 (2015).
Jin, Y. & Hammell, M. Analysis of RNA-seq data using TEtranscripts. Methods Mol. Biol. 1751, 153–167 (2018).
Tokuyama, M. et al. ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc. Natl Acad. Sci. USA 115, 12565–12572 (2018).
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
Acknowledgements
We thank A. Iwasaki and M. Tokuyama for help with the ERV transcriptome analysis. We thank members of the Bass laboratory, S. Kitajima, S. Gu, X. Wang and H. Singh, for insightful discussions. We thank D.E. Ingber for kindly sharing the influenza A/WSN/33 (H1N1) virus strain and influenza A/PR8/34 (H1N1) virus strain, M. Berkeley and the MBCF Core for assistance with RNA sequencing, and the Molecular Pathology Core of P01 CA098101 for assistance with immunohistochemistry staining. The research was supported by the Twomey Family Fellowship in Esophageal Cancer Research (J.Z.). A.J.B. was supported by NIH grants no. R01 CA196932 and no. R01 CA187119. A.K.R., K.-K.W. and A.J.B. were supported by NIH grant no. P01 CA098101. X.Z. was supported by NIH grant no. R00 CA215244.
Author information
Authors and Affiliations
Contributions
Z.W., J.Z., X.Z. and A.J.B. conceived the project. Z.W., J.Z., X.Z., J.B.L., P.C., F.G.A., A.K.N., L.B.G., L.S., K.L., Yanli Liu., Y.Z., J.Y.B. and J.H. performed the experiments. Z.W., X.Z., Z.Z., Y.X., Z.V.H., A.P., X.Q., J.M.M., N.K., Yang Liu and C.B. performed the computational analyses and statistical calculations. F.G.A., M.S. and D.T.P. viewed the pathology slides. I.C., L.S., D.T.P., J.P.K., J.H., J.Q., J.J.I., H.N., A.K.R., K.-K.W., M.M., D.A.B., M.B. and H.L. helped with the resources. A.K.R., K.-K.W., M.M., D.A.B., M.B., H.L. and A.J.B. provided supervision. Z.W., J.Z., X.Z. and A.J.B. wrote the manuscript with help from all the authors. Z.Z., Y.X. and J.B.L. contributed equally.
Corresponding author
Ethics declarations
Competing interests
A.J.B. receives research funding from Bayer, Merck and Novartis; is a consultant to Earli and HelixNano; and is a cofounder of Signet Therapeutics. K.-K.W. is a founder and equity holder of G1 Therapeutics and has consulting/sponsored research agreements with MedImmune, Takeda, TargImmune, BMS, AstraZeneca, Janssen, Pfizer, Novartis, Merck, Ono and Array. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks David Fernandez-Antoran, Andrew Sharrocks and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Overexpression of Sox2 and loss of Trp53/Cdkn2a give rise to esophageal squamous cell carcinoma (ESCC).
a, Immunoblots and quantification of p63 expression in four engineered organoids (normalized to β-actin) (representative image from 2 independent experiments). b, Immunoblots of Sox2 in mouse organoids, human ESCC cell lines and a normal esophageal epithelium cell line EPC2 (representative image from 2 independent experiments).
Extended Data Fig. 2 Genomic occupancy of Sox2 changes from normal to neoplastic organoids.
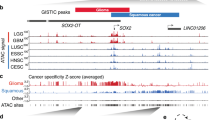
a, Pairwise Spearman correlation of Sox2 ChIP-seq between the Sox2 overexpression and non-overexpression organoids. Unsupervised hierarchical clustering showed the relatedness of each sample. Two independent biological replicates were performed and shown for each organoid. b, Pairwise Spearman correlation of ATAC-seq between the Sox2 overexpression and non-overexpression organoids. Unsupervised hierarchical clustering shows the correlation for each pairs of the biological replicates. Two independent biological replicates were performed and shown for each organoid. c, Pairwise Spearman correlation of H3K27ac ChIP-seq between the Sox2 overexpression and non-overexpression organoids. Unsupervised hierarchical clustering showed the correlation between each sample pair from the same organoid. Two independent biological replicates were performed and shown for each organoid. d, Heatmaps of ATAC-seq signals ±2 kb around Sox2 peaks (identified in Fig. 2a) across different organoids. Two independent biological replicates were performed and shown for each organoid. e, Venn Diagram shows the overlap of Sox2 sites that are gained in SCPP (vs. Normal) and open chromatin sites in SCPP (vs. Normal), as defined by ATAC-seq signal. P < 0.0001 calculated by two-sided fisher exact test (SCPP vs. Normal (unchanged chromatin sites as control). f, Representative ATAC-seq tracks, showing increased chromatin accessibility at Klf5, Klf12, Stat3, Mavs, Ifih1 and Fbxo47 locus in normal, SC and SCPP organoids.
Extended Data Fig. 3 Evolution of genomic occupancy and Sox2 activation changes from normal to neoplastic organoids.
a, Heatmaps of H3K27ac ChIP-seq signal ±2 kb around Sox2 peaks (identified in Fig. 2a) across different organoids. Two independent biological replicates were performed and shown for each organoid. b, H3K4me1 ChIP-seq signals ±2 kb around Sox2 peaks (identified in Fig. 2a) across different organoids. Two independent biological replicates were performed and shown for each organoid. c, The ratio of SOX2 positive correlated genes from TCGA ESCC in Sox2 up/down/unaffected genes in the comparison of SCPP vs Normal organoid models, **** p < 0.0001 calculated by two-sided fisher exact test for 2 × 2 contingency table. d, Genes that are significantly positively correlated with SOX2 in TCGA ESSC samples as a ‘Gene Set’ with Gene Set Enrichment Analysis (GSEA) used to quantify expression differences of the gene set between SCPP and Normal organoids. (Two-sided FDR-adjusted q-value=0.106; nominal p = 0.106 calculated by the GSEA package) e, Genes that are significantly positively correlated with SOX2 in TCGA ESSC samples as a ‘Gene Set’ with GSEA is used to quantify expression differences of the gene set between SCPP and CPP organoids. (Two-sided FDR-adjusted q-value<0.0001; nominal p < 0.0001 calculated by the GSEA package).
Extended Data Fig. 4 Sox2 overexpression creates new super-enhancers (SE) and activates cancer promoting genes.
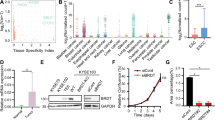
a, The percentage of super-enhancers harboring gained Sox2 binding sites (left) or gained chromatin open sites (right). The super-enhancers are categorized as ‘Gained’, ‘Shared’ and ‘Lost’ based on the comparison between SCPP and Normal organoids. Gained Sox2 sites or chromatin open sites refer to the ones gained in SCPP organoids compared to Normal organoids. b, Venn diagram showing overlap of gained SEs with Sox2 binding sites gained in SCPP (vs. Normal) and gained SEs with open chromatin sites, defined by ATAC-seq in SCPP (vs. Normal). c, Venn diagram showing overlap of gained SEs in SCPP (vs Normal) and the Cancer Gene Census. d, mRNA expression levels of Il6ra and Stat3 mRNA in 4 groups organoids by quantitative RT-PCR. Data are shown as Mean ± SD. NS: not significant and p-value calculated by two-sided unpaired t-tests. Three independent biological replicates were performed for each organoid. e, Immunoblot of p-Stat3 and total Stat3 in different organoids (representative image from 3 independent experiments) (up), and the quantification of the p-Stat3 expression in different organoids (down). f, Venn Diagram shows the overlap of super-enhancers that are gained in SCPP (vs. Normal) and shared super-enhancers across three ESCC cell lines (TT, TE10 and KYSE70). P < 0.0001 calculated by two-sided fisher exact test (ESCC super-enhancer unrelated genes as control). g, Representative SOX2 and H3K27ac ChIP-seq tracks showing super-enhancers at STAT3 and IL6R locus in ESCC cell lines TE10 and TT. h, Immunoblot of p-STAT3 and total STAT3 in ESCC cell line TE10 and TT after shRNA-mediated SOX2 silencing (representative image from 2 independent experiments).
Extended Data Fig. 5 Sox2 overexpression activates Klf5 by binding to Klf5 enhancer.
a, Sox2 and H3K27ac ChIP-seq and ATAC-seq profiles in Normal and SCPP organoids identify constituent enhancers e1-e3. b, Luciferase reporter assays measuring the enhancer activity of e1-e3 in four groups of organoids. The pGL3 plasmid without the enhancer region (empty) is used as a negative control. Along the Y-axis, relative Luciferase units are normalized to negative control. Three biological replicates were performed for each organoid. Data are shown as Mean ± SD, p-value calculated by one-way ANOVA followed by Benjamini-Hochberg correction. c, Luciferase reporter assays measuring the enhancer activity of E3 with and without the Sox2 motif deleted in SCPP organoids. The pGL3 plasmid without the enhancer region (empty) is used as a negative control. Along the Y-axis, relative Luciferase units are normalized to negative control. Four biological replicates were performed for each organoid. Data are shown as Mean ± SD, p-value calculated by two-sided t-tests. d, mRNA expression of Klf5 in SCPP organoids with and without dCAS9-mediated e1-e3 enhancer repression as measured by RT-PCR. The sgNT is used as a negative control. Four biological replicates were performed. Data are shown as Mean ± SD, p-value calculated by one-way ANOVA followed by Benjamini-Hochberg correction.
Extended Data Fig. 6 KLF5 facilitates enhanced SOX2 activity in squamous tumorigenesis.
a, SOX2-KLF5 interaction, shown by co-IP of SOX2 (top) and KLF5 (bottom), followed by immunoblotting in human ESCC cell line TE10, TT and KYSE70. b, Overlap of binding sites gained in SCPP (vs. Normal) by Sox2 and Klf5 ChIP-seq. P < 0.0001; two-sided fisher exact test with unchanged Klf5 binding sites as control. c, Percentage of gained super-enhancers in SCPP (from Fig. 3b) with Klf5 ChIP-seq occupancy. d, Immunoblots of Klf5 and Sox2 after shRNA-mediated Sox2 silencing in SCPP (representative image from 2 independent experiments). e, Overlap of downregulated genes upon shSOX2 and shKLF5 in KYSE70 (Left) and TT (Right). P < 0.0001; two-sided fisher exact test with shKLF5 unchanged genes as control. f, Genes that are significant down-regulated by shKLF5 in ESCC cell lines as a ‘Gene Set’ with GSEA to quantify their expression differences with shSOX2 in KYSE70 (Left) and TT (Right) with two-sided FDR-adjusted q-value<0.0001; nominal p < 0.0001 calculated using the GSEA package. g, Differential expression level of 95 genes regulated by SOX2 and KLF5 (genes down-regulated by shSOX2 and shKLF5 in KYSE70 and TT) in TCGA ESCC (Esophageal squamous cell carcinoma) EAC (Esophageal adenocarcinoma) and Normal esophagus samples. P < 0.0001 **** by two-sided t-tests. Boxes indicate interquartile range, and whiskers show maximum and minimum values. Horizontal line, median. h, Genes regulated by SOX2 and KLF5 in ESCC cell lines as a ‘Gene Set’ with GSEA to measure their expression differences between ESCC vs EAC (Top) and ESCC vs Normal (Bottom) with two-sided FDR-adjusted q-value<0.0001; nominal p < 0.0001 calculated using the GSEA package. i, Overlap between SOX2/KLF5 putative target genes and significantly downregulated genes upon shSOX2 (left) and shKLF5 (right) in KYSE70 (top) and TT (bottom). SOX2/KLF5 target genes are defined by intersection of 1) Significantly upregulated genes in SCPP vs Normal; 2) Genes that gain Sox2 and Klf5 binding sites in SCPP vs Normal. P-value; two-sided fisher exact test with SOX2/KLF5 unrelated genes as control.
Extended Data Fig. 7 Klf5 facilitates SOX2 activity and is essential for SOX2 + ESCC.
a, ATAC-seq signal (horizontal window of ±2 kb from the peak center) in open-chromatin sites lost upon shSOX2 in KYSE70 (Top). (Bottom) Overlap of open-chromatin sites lost upon shSOX2 and endogenous SOX2 binding sites in KYSE70 cells. P < 0.0001 by fisher exact test with KYSE70 shSOX2 unchanged chromatin sites as control. b, ATAC-seq signal in open-chromatin sites gained and lost upon shKLF5 in KYSE70 (Top). Two independent biological replicates are shown. (Bottom) overlap of open-chromatin sites lost upon shKLF5 and endogenous SOX2 binding sites in KYSE70. P < 0.0001 by fisher exact test with KYSE70 shKLF5 unchanged chromatin sites as control. c, Overlap of lost ATAC-seq signals after shRNA-mediated SOX2 or KLF5 silencing in KYSE70. P < 0.0001 by two-sided fisher exact test (KYSE70 shKLF5 unchanged sites as control). d, Tumor formation rate of SCPP organoids with or without Klf5 silencing. Each group includes 6–10 tumors. Ns: not significant, p-value by two-sided fisher exact test. e, Immunoblots following siKLF5 in human ESCC cell lines. f, Proliferation TE10, KYSE70 and TT after KLF5 siRNA. Three independent biological replicates were performed for each cell line. Data were normalized to control siRNA and shown as mean ± SD, p-value by one-way ANOVA with Benjamini-Hochberg correction. g, Representative immunoblots of KYSE70 72 hours after shRNA-mediated SOX2 and KLF5 silencing. h, Representative immunoblots of TT 72 hours after shRNA-mediated SOX2 and KLF5 silencing. i, Proliferation of KYSE70 (Top) and TT (Bottom) 72 hours after shRNA-mediated SOX2 and KLF5 silencing. Two biological replicates were performed. j, Growth for KYSE70 xenografts (n = 6–10) with shKLF5. Data are shown as mean ± SEM; ****p < 0.0001 by two-sided two-way ANOVA with Benjamini-Hochberg correction. k, Growth for KYSE70 xenografts (n = 6–10) with shSOX2. Data are shown as mean ± SEM; ****p < 0.0001 by two-sided two-way ANOVA with Benjamini-Hochberg correction. l, Tumor volume of KYSE70 with noted inducible shRNA. Representative images of tumors from panel. Scale bar is 2 cm. 6–10 tumors per group.
Extended Data Fig. 8 SOX2 overexpression induces IFN activation and promotes ADAR1 dependency.
a-c, ADAR1 dependency from genome-wide CRISPR screening in (a) 608 SOX2 amplified cell lines versus 16 non SOX2 amplified cell lines (b) 65 SCCs versus 246 adenocarcinoma cell lines (c) 10 SOX2 amplified SCC versus 55 non SOX2 amplified SCC cell lines. Box and whisker plots show the median (center line), interquartile range (box), 1.5 x interquartile range (whiskers), and outliers (points). Two-sided Mann-Whitney U tests were performed to assess significant differences in ADAR1 dependency, which are defined using CERES scores from the Dependency Map’s 19Q3 data release. SOX2 amplified lines are defined as those with log2(copy ratio) > 1. d, Representative images of crystal violet staining from noted human ESCC cell lines grown for two weeks treatment with or without doxycycline induction of indicated ADAR1 shRNA. Two biological replicates were performed. e, Tumor volume of 2 × 106 KYSE70 cells transduced with indicated inducible ADAR1 shRNA injected into flanks of nude mice and provided doxycycline-containing food (625 ppm). Representative images of tumors from panel. Scale bar is 2 cm. 4–8 tumors per group. f, Immunoblots of Adar1 depletion in CPP and SCPP organoids following transduction with indicated Adar1 sgRNA and with transfection of control, dsRNA (poly IC) and dsDNA (poly dAdT). (This experiment was repeated once with similar results). g, mRNA expression of Mx1, Cxcl10 and Isg15 in CPP and SCPP by quantitative-PCR. Three biological replicates were performed for each organoid. Data are shown as mean ± SD, p-value calculated by two-sided unpaired t-tests. h, Gene-Set Enrichment Analysis (GSEA) of pathways upregulated in SCPP compared with CPP organoids. (FDR-adjusted q-value calculated using the GSEA package) i, Enrichment scores (ES) plots for the interferon alpha and gamma gene sets in comparison of mRNA profiles of SCPP compared to CPP organoids. (Left, two-sided FDR-adjusted q-value=0.024; nominal p = 0.008 calculated using the GSEA package) (Right, two-sided FDR-adjusted q-value=0.015; nominal p = 0.034 calculated using the GSEA package) j, The most statistically enriched pathway identified from significant upregulation gained super-enhancer related genes in the comparison of SCPP vs Normal organoids (nominal p-value calculated using the GSEA package).
Extended Data Fig. 9 Sox2/Klf5 complexes induce IFN activation.
a, Most enriched pathways from upregulated genes at loci of gained Sox2 binding sites (SCPP vs Normal organoids); nominal p-value from GSEA package. b, Representative ATAC-seq, Sox2 and H3K27ac ChIP-seq tracks, showing increased accessibility, Sox2 binding and H3K27ac level at Tmem173 (Sting), Mavs and Dhx58 (Lgp2) loci in SCPP (versus CPP). c, mRNA expression of Sting and Lgp2 Normal and SCPP by mRNA seq; two biological replicates performed. d, Quantification of binding of Sox2 and Klf5 as assessed by ChIP-PCR at loci of Tmem173 (Sting), Il6ra and Ifih1(Mda5); Three biological replicates. Data are shown as mean ± SD; p-value by two-sided t-tests. e, mRNA expression Mx1, Cxcl10 and Isg15 in CPP and SCPP as assessed by quantitative RT-PCR 24 h after infection with H1N1 flu virus; tree independent biological replicates. Data are shown as mean ± SD, p-value calculated by two-sided unpaired t-tests. f, mRNA expression of Mx1, Cxcl10 and Isg15 in CPP and SCPP as assessed by quantitative RT-PCR 24 h after transfection with 5’ppp-dsRNA (normalized to 5’ppp-dsRNA control); three independent biological replicates. Data are shown as mean ± SD, NS: not significant as calculated by two-sided unpaired t-tests. g, Quantification of J2(dsRNA) and Adar1 staining in CPP and SCPP (Image J software) with signals normalized to cell number and then CPP; three technical replicates per condition. The data are shown as mean ± SD, p-value calculated by two-sided unpaired t-tests. h, Representative double immunofluorescence images for J2(dsRNA; green) and Adar1 (red) and DAPI (blue) in SCPP with Adar1 silencing. Scale bar=40um. (This experiment was repeated once with similar results). i, Quantification of J2(dsRNA) (Left) and Adar1 (Right) in SCPP with CRISPR-mediated Adar1 knockout. The signal of sgAdar1 was normalized to cell number and then sgNT; three technical replicates. The data are shown as mean ± SD, p-value by one-way ANOVA test with Benjamini-Hochberg correction.
Extended Data Fig. 10 SOX2 enhances dsRNA expression by activating endogenous retroviruses transcription.
a, Pie chart depicts the number of upregulated ERV families in SCPP versus CPP with/without enrichment of gained Klf5 binding sites. b, Bar graph illustrates the enrichment of gained Klf5-binding sites in specific ERV families highlighted in Supplemental Fig. 6a. The background ratio was determined by Klf5 ChIP-seq gained sites/ unaffected sites in SCPP vs CPP. P-value calculated by two-sided fisher exact test (Background as control). c, Representative H3K27ac ChIP-seq, Sox2 ChIP-seq, Klf5 ChIP-seq and ATAC-seq tracks, showing increased H3K27ac level, gained Sox2 binding, gained Klf5 binding and increased chromatin accessibility at different RLTR13D6 loci in SCPP versus CPP organoids. d, The relative expression of representative RLTR13D6 ERVs in different organoids was assessed by quantitative RT-PCR. Three independent biological replicates were performed for each condition. Data are shown as mean ± SD, p-value calculated by two-sided t-tests. e, The relative ERV expressions of different gained H3K27ac sites (SCPP vs CPP) across four organoids by quantitative PCR. Three biological replicates were performed for each organoid. The data are shown as mean ± SD, p-value calculated by two-sided t-tests. f, Representative double immunofluorescence images for dsRNA marker J2(dsRNA) and Adar1 in SCPP organoid with shRNA-mediated KLF5 silencing. FITC (green), phalloidin (red) and DAPI (blue) channels were used to detect J2(dsRNA), Adar1, and nucleus, respectively. Scale bar=40 um. (This experiment was repeated once with similar results). g. Quantification of the J2(dsRNA) staining in SCPP organoid with shRNA-mediated Klf5 silencing. The signal of shKlf5 was normalized to cell number and then shNT. Three technical replicates were performed for each condition. The data are shown as mean ± SD, NS: not significant and ****p < 0.0001 as calculated by the two-sided one-way ANOVA test followed by Benjamini-Hochberg correction. h, Quantification of the J2(dsRNA) staining in ESCC cell line TT and TE10 with shRNA-mediated SOX2 or KLF5 knockdown. Data are normalized to cell number and Dox (-) and shown as mean ± SD, NS: not significant and ****p < 0.0001 as calculated by two-sided unpaired t-tests.
Supplementary information
Supplementary Information
Supplementary Methods
Supplementary Tables 1–8
Supplementary Table 1. Mouse WES SNP_annotation. Supplementary Table 2. Normal SCPP super-enhancer. Supplementary Table 3. TT KYSE70 shSOX2 shKLF5 overlap genes. Supplementary Table 4. Mouse organoids Sox2 Klf5 coregulated genes. Supplementary Table 5. Sox2 upregulated mouse genes contain 3ʹ UTR ERV. Supplementary Table 6. TT KYSE70 shSOX2 shKLF5 downregulated ERVs. Supplementary Table 7. Primer information. Supplementary Table 8. DepMap sample annotation.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Wu, Z., Zhou, J., Zhang, X. et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat Genet 53, 881–894 (2021). https://doi.org/10.1038/s41588-021-00859-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00859-2
This article is cited by
-
RNA editing enzymes: structure, biological functions and applications
Cell & Bioscience (2024)
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization
Cell Death & Differentiation (2024)
-
GSK3β-driven SOX2 overexpression is a targetable vulnerability in esophageal squamous cell carcinoma
Oncogene (2023)
-
Esophageal organoids: applications and future prospects
Journal of Molecular Medicine (2023)
-
ADAR1 polymorphisms contribute to increased susceptibility in pediatric acute lymphoblastic leukemia
Annals of Hematology (2023)