Abstract
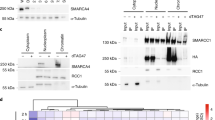
Aberrations in genes coding for subunits of the BRG1/BRM associated factor (BAF) chromatin remodeling complexes are highly abundant in human cancers. Currently, it is not understood how these mostly loss-of-function mutations contribute to cancer development and how they can be targeted therapeutically. The cancer-type-specific occurrence patterns of certain subunit mutations suggest subunit-specific effects on BAF complex function, possibly by the formation of aberrant residual complexes. Here, we systematically characterize the effects of individual subunit loss on complex composition, chromatin accessibility and gene expression in a panel of knockout cell lines deficient for 22 BAF subunits. We observe strong, specific and sometimes discordant alterations dependent on the targeted subunit and show that these explain intracomplex codependencies, including the synthetic lethal interactions SMARCA4–ARID2, SMARCA4–ACTB and SMARCC1–SMARCC2. These data provide insights into the role of different BAF subcomplexes in genome-wide chromatin organization and suggest approaches to therapeutically target BAF-mutant cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Next-generation sequencing data have been deposited with the NCBI GEO (GSE108390). Mass spectrometry data have been deposited with the PRIDE archive PXD013102. The processed data used for the analyses are available at http://baf-complex.computational-epigenetics.org.
Code availability
Software used for the analyses is available at http://baf-complex.computational-epigenetics.org.
References
Wang, W. et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15, 5370–5382 (1996).
Kadoch, C. & Crabtree, G. R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 (2015).
Phelan, M. L., Sif, S., Narlikar, G. J. & Kingston, R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253 (1999).
Chandler, R. L. et al. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell Biol. 33, 265–280 (2013).
Hodges, C., Kirkland, J. G. & Crabtree, G. R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 (2016).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Hoffman, G. R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl Acad. Sci. USA 111, 3128–3133 (2014).
Wilson, B. G. et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol. Cell. Biol. 34, 1136–1144 (2014).
Oike, T. et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 73, 5508–5518 (2013).
Helming, K. C. et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 20, 251–254 (2014).
Kelso, T. W R. et al. Chromatin accessibility underlies synthetic lethality of SWI/SNFsubunits in ARID1A-mutant cancers. eLife 6, e30506 (2017).
Mathur, R. et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017).
Mashtalir, N. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288.e20 (2018).
Alpsoy, A. & Dykhuizen, E. C. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 293, 3892–3903 (2018).
Dutta, A. et al. Composition and function of mutant Swi/Snf Complexes. Cell Rep. 18, 2124–2134 (2017).
Sen, P. et al. Loss of Snf5 induces formation of an aberrant SWI/SNF complex. Cell Rep. 18, 2135–2147 (2017).
Blomen, V. A. et al. Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015).
Yan, Z. et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19, 1662–1667 (2005).
Michel, B. C. et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410–1420 (2018).
Pan, J. et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 6, 555–568.e7 (2018).
Wang, X. F. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Hodges, H. C. et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 25, 61–72 (2018).
Raab, J. R., Resnick, S. & Magnuson, T. Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 11, e1005748 (2015).
Valianou, M. et al. Pharmacological inhibition of Polo-like kinase 1 (PLK1) by BI-2536 decreases the viability and survival of hamartin and tuberin deficient cells via induction of apoptosis and attenuation of autophagy. Cell Cycle 14, 399–407 (2015).
Wang, N. N. et al. Molecular targeting of the oncoprotein PLK1 in pediatric acute myeloid leukemia: RO3280, a novel PLK1 inhibitor, induces apoptosis in leukemia cells. Int. J. Mol. Sci. 16, 1266–1292 (2015).
Zhao, C. L. et al. Downregulation of PLK1 by RNAi attenuates the tumorigenicity of esophageal squamous cell carcinoma cells via promoting apoptosis and inhibiting angiogenesis. Neoplasma 62, 748–755 (2015).
Wang, W. et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130 (1996).
Zhao, K. et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1998).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018).
Stransky, N. et al. Pharmacogenomic agreement between two cancer cell line data sets. Nature 528, 84–87 (2015).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
McDonald, E. R.3rd et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592.e10 (2017).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 1–9 (2015).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Jiang, H., Lei, R., Ding, S. W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Quinlan, A. R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 1–34 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Hoffman, M. M. et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 41, 827–841 (2013).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Glaus, P., Honkela, A. & Rattray, M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics 28, 1721–1728 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Sheffield, N. C. & Bock, C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 32, 587–589 (2016).
Sheffield, N. C. et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 23, 777–788 (2013).
Sánchez-Castillo, M. et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 43, D1117–D1123 (2015).
Rosenbloom, K. R. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681 (2015).
Liu, T. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011).
Adams, D. et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 30, 224–226 (2012).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Schep, A. N. et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015).
Acknowledgements
We thank the Biomedical Sequencing Facility, the Proteomics and Metabolomics Facility and the Platform Austria for Chemical Biology at CeMM for their support in generating and analyzing the next-generation sequencing, proteomics or screening data, respectively. We gratefully acknowledge Horizon Discovery for providing the HAP1 cell lines, and Boehringer Ingelheim, the Superti-Furga laboratory (CeMM) and Winter laboratory (CeMM) for providing various cancer cell lines. We acknowledge the experimental support provided by J. Block and D. Donertas. Research in the Kubicek laboratory is supported by the Austrian Federal Ministry for Digital and Economic Affairs and the National Foundation for Research, Technology and Development, the Austrian Science Fund (FWF) F4701 and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-CoG-772437). C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant no. 679146).
Author information
Authors and Affiliations
Contributions
S.S., G.B., M.P. and S.K. planned the study and designed the experiments. S.S., K.R., M.H., T.P., K.P., C.S., A.R. and B.B. performed the experiments. A.F.R., P.M., L.V., S.S. and S.K. analyzed the data. S.S. and S.K. wrote the manuscript. S.K., M.P., G.B., A.C.M., C.B. and J.M. supervised the work. S.K. provided the funding.
Corresponding author
Ethics declarations
Competing interests
G.B. and M.P. are employees of Boehringer Ingelheim RCV GmbH & Co KG.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and Supplementary Note
Supplementary Table 1
Materials.
Supplementary Table 2
ATAC-seq.
Supplementary Table 3
ChIP-seq.
Supplementary Table 4
RNA-seq.
Supplementary Table 5
Synthetic lethality.
Supplementary Table 6
Statistics.
Rights and permissions
About this article
Cite this article
Schick, S., Rendeiro, A.F., Runggatscher, K. et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat Genet 51, 1399–1410 (2019). https://doi.org/10.1038/s41588-019-0477-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0477-9
This article is cited by
-
Context-specific functions of chromatin remodellers in development and disease
Nature Reviews Genetics (2024)
-
Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility
Nature Genetics (2024)
-
Tracking chromatin state changes using nanoscale photo-proximity labelling
Nature (2023)
-
Neurodevelopmental disorders, like cancer, are connected to impaired chromatin remodelers, PI3K/mTOR, and PAK1-regulated MAPK
Biophysical Reviews (2023)
-
SWI/SNF Chromatin Remodelers: Structural, Functional and Mechanistic Implications
Cell Biochemistry and Biophysics (2023)