Abstract
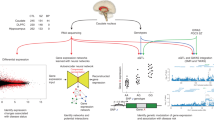
Transcriptomic imputation approaches combine eQTL reference panels with large-scale genotype data in order to test associations between disease and gene expression. These genic associations could elucidate signals in complex genome-wide association study (GWAS) loci and may disentangle the role of different tissues in disease development. We used the largest eQTL reference panel for the dorso-lateral prefrontal cortex (DLPFC) to create a set of gene expression predictors and demonstrate their utility. We applied DLPFC and 12 GTEx-brain predictors to 40,299 schizophrenia cases and 65,264 matched controls for a large transcriptomic imputation study of schizophrenia. We identified 413 genic associations across 13 brain regions. Stepwise conditioning identified 67 non-MHC genes, of which 14 did not fall within previous GWAS loci. We identified 36 significantly enriched pathways, including hexosaminidase-A deficiency, and multiple porphyric disorder pathways. We investigated developmental expression patterns among the 67 non-MHC genes and identified specific groups of pre- and postnatal expression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Our CMC-derived DLPFC prediction models are publicly available at https://github.com/laurahuckins/CMC_DLPFC_prediXcan.
Change history
13 May 2019
In the HTML version of the article originally published, the author group ‘The Schizophrenia Working Group of the Psychiatric Genomics Consortium’ was displayed incorrectly. The error has been corrected in the HTML version of the article.
References
Nicolae, D. L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Dobbyn, A. et al. Co-localization of conditional eQTL and GWAS signatures in schizophrenia. Preprint at https://www.biorxiv.org/content/10.1101/129429v2 (2017).
Gilad, Y., Rifkin, S. A. & Pritchard, J. K. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24, 408–415 (2008).
Cookson, W., Liang, L., Abecasis, G., Moffatt, M. & Lathrop, M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10, 184–194 (2009).
Albert, F. W. & Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16, 197–212 (2015).
Moffatt, M. F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007).
Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Dubois, P. C. A. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Libioulle, C. et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 3, e58 (2007).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Boocock, J., Giambartolomei, C. & Stahl, E. A. COLOC2 (2016).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Geschwind, D. H. & Flint, J. Genetics and genomics of psychiatric disease. Science 349, 1489–94 (2015).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Bennett, D. A., Schneider, J. A., Arvanitakis, Z. & Wilson, R. S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 9, 628–645 (2012).
Bennett, D. A., Schneider, J. A., Buchman, A. S., Barnes, L. L. & Wilson, R. S. Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 9, 646–663 (2012).
Mele, M. et al. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Dobbyn, A. et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and Co-localization with schizophrenia GWAS. Am. J. Hum. Genet. https://doi.org/10.1016/j.ajhg.2018.04.011(2018).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Benjamin, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Pardiñas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Sanders, S. J. First glimpses of the neurobiology of autism spectrum disorder. Curr. Opin. Genet. Dev. 33, 80–92 (2015).
Monkol, Lek. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Malhotra, D. et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron 72, 951–963 (2011).
Bautista, O., Vázquez-Caubet, J. C., Zhivago, E. A. & Dolores Sáiz, M. From metabolism to psychiatric symptoms: psychosis as a manifestation of acute intermittent porphyria. J. Neuropsychiatry Clin. Neurosci. 26, E30 (2014).
Zimmermann, M., Bonaccurso, C., Valerius, C. & Hamann, G. F. Acute intermittent porphyria. A clinical chameleon: case study of a 40-year-old female patient. Nervenarzt 77, 1501–1505 (2006).
Ventura, P. et al. A challenging diagnosis for potential fatal diseases: recommendations for diagnosing acute porphyrias. Eur. J. Intern. Med. 25, 497–505 (2014).
Pischik, E. & Kauppinen, R. An update of clinical management of acute intermittent porphyria. Appl. Clin. Genet. 8, 201–214 (2015).
Kumar, B. Acute intermittent porphyria presenting solely with psychosis: a case report and discussion. Psychosomatics 53, 494–498 (2012).
Bonnot, O. et al. Diagnostic and treatment implications of psychosis secondary to treatable metabolic disorders in adults: a systematic review. Orphanet J. Rare Dis. 9, 65 (2014).
Kaback, M. M. & Desnick, R. J. Hexosaminidase A Deficiency: GeneReviews (University of Washington, Seattle, 1993).
Osama, S. Late onset Tay-Sachs disease presenting as a brief psychotic disorder with catatonia: a case report and review of literature. Jefferson J. Psych. 15, 4 (2000).
Skaper, S. D. in Brain Protection in Schizophrenia, Mood and Cognitive Disorders (ed. Ritsner, M. S.) 135–165 (Springer Science & Business Media, 2010).
Castellano, E. et al. RAS signalling through PI3-Kinase controls cell migration via modulation of Reelin expression. Nat. Commun. 7, 11245 (2016).
Gururajan, A. & Buuse, M. van den. Is the mTOR-signalling cascade disrupted in Schizophrenia? J. Neurochem. 129, 377–387 (2014).
Ritsner, M. S. Brain Protection in Schizophrenia, Mood and Cognitive Disorders (Springer Science & Business Media, 2010).
Enriquez-Barreto, L. & Morales, M. The PI3K signaling pathway as a pharmacological target in Autism related disorders and Schizophrenia. Mol. Cell. Ther. 4, 2 (2016).
Glessner, J. T. et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc. Natl Acad. Sci. USA 107, 10584–10589 (2010).
Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 18, 199–209 (2015).
Bauman, A. L. et al. Cocaine and antidepressant-sensitive biogenic amine transporters exist in regulated complexes with protein phosphatase 2A. J. Neurosci. 20, 7571–7578 (2000).
Ayadi, A. et al. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm. Genome 23, 600–610 (2012).
Keane, T. M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011).
Howe, D. G. et al. ZFIN, the zebrafish model organism database: increased support for mutants and transgenics. Nucleic Acids Res. 41, D854–D860 (2013).
Smith, C. L., Blake, J. A., Kadin, J. A., Richardson, J. E. & Bult, C. J. Mouse genome database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 46, D836–D842 (2018).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Nguyen, H. T. et al. Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med. 9, 114 (2017).
Mancuso, N. et al. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 100, 473–487 (2017).
Gottlieb, A., Daneshjou, R., DeGorter, M., Montgomery, S. & Altman, R. Population-specific imputation of gene expression improves prediction of pharmacogenomic traits for African Americans. Preprint at https://www.biorxiv.org/content/10.1101/115451v1 (2017).
Need, A. & Goldstein, D. B. Next generation disparities in human genomics: concerns and remedies. Trends Genet 25, 489–494 (2009).
Popejoy, A. & Fullerton, S. Genomics is failing on diversity. Nature 538, 161–164 (2016).
Pardiñas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and maintained by background selection. Preprint at https://www.biorxiv.org/content/10.1101/068593v1 (2016).
Browning, R. in The Poems of Robert Browning (eds Porter, C. & Clarke, H. A.) 257–271 (Thomas Y. Cromwell and Company, 1896).
Loftus, L. S. & Arnold, W. N. Vincent van Gogh’s illness: acute intermittent porphyria? BMJ 303, 1589–1591 (1991).
Strik, W. K. The psychiatric illness of Vincent van Gogh. Nervenarzt 68, 401–409 (1997).
Arnold, W. N. The illness of Vincent van Gogh. J. Hist. Neurosci. 13, 22–43 (2004).
Hughes, J. R. A reappraisal of the possible seizures of Vincent van Gogh. Epilepsy Behav. 6, 504–510 (2005).
Bhattacharyya, K. B. & Rai, S. The neuropsychiatric ailment of Vincent van Gogh. Ann. Indian Acad. Neurol. 18, 6–9 (2014).
Correa, R. Vincent van Gogh: A pathographic analysis. Med. Hypotheses 82, 141–144 (2014).
Peters, T. J. & Beveridge, A. The madness of King George III: a psychiatric re-assessment. Hist. Psychiatry 21, 20–37 (2010).
Szatkiewicz, J. P. et al. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry 19, 762–773 (2014).
Fromer, M. et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Keefe, R. S. E. & Fenton, W. S. How should DSM-V criteria for schizophrenia include cognitive impairment? Schizophr. Bull. 33, 912–920 (2007).
Reichenberg, A. et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am. J. Psychiatry 167, 160–169 (2010).
Gold, J. M. Cognitive deficits as treatment targets in schizophrenia. Schizophr. Res. 72, 21–28 (2004).
Cannon, M. et al. Evidence for early-childhood, pan-developmental impairment specific to schizophreniform disorder. Arch. Gen. Psychiatry 59, 449 (2002).
Parikshak, N. N., Gandal, M. J., Geschwind, D. H. & Angeles, L. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458 (2015).
Glass, D. et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 14, R75 (2013).
Colantuoni, C. et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2012).
Gusev, A. et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Preprint at https://www.biorxiv.org/content/10.1101/067355v1 (2016).
Ardlie, K. G. et al. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
COCHRAN, W. G. THE comparison of percentages in matched samplES. Biometrika 37, 256–266 (1950).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Kirov, G. et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 17, 142–153 (2012).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25–29 (2000).
The Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2014).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Croft, D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 (2014).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Lin, G. N. et al. Spatiotemporal 16p11.2 Protein network implicates cortical late mid-fetal brain development and KCTD13- Cul3-RhoA pathway in psychiatric diseases. Neuron 85, 742–754 (2015).
Bahl, E., Koomar, T. & Michaelson, J. J. cerebroViz: An R package for anatomical visualization of spatiotemporal brain data. Bioinformatics 33, btw726 (2016).
van der Weyden, L., White, J. K., Adams, D. J. & Logan, D. W. The mouse genetics toolkit: revealing function and mechanism. Genome Biol. 12, 224 (2011).
Brown, S. D. M. & Moore, M. W. The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–640 (2012).
Acknowledgements
We dedicate this manuscript to the memory of Pamela Sklar, whose guidance and wisdom we miss daily. We strive to continue her legacy of thoughtful, innovative, and collaborative science. Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692.
Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: P. Sklar, J. Buxbaum (Icahn School of Medicine at Mount Sinai), B. Devlin, D. Lewis (University of Pittsburgh), R. Gur, C.-G. Hahn (University of Pennsylvania), K. Hirai, H. Toyoshiba (Takeda Pharmaceuticals Company Limited), E. Domenici, L. Essioux (F. Hoffman-La Roche Ltd), L. Mangravite, M. Peters (Sage Bionetworks), T. Lehner, B. Lipska (NIMH).
ROSMAP study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute.
The iPSYCH-GEMS team acknowledges funding from the Lundbeck Foundation (grant no. R102-A9118 and R155-2014-1724), the Stanley Medical Research Institute, an Advanced Grant from the European Research Council (project no. 294838), the Danish Strategic Research Council the Novo Nordisk Foundation for supporting the Danish National Biobank resource, and grants from Aarhus and Copenhagen Universities and University Hospitals, including support to the iSEQ Center, the GenomeDK HPC facility, and the CIRRAU Center.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on September 5, 2016. BrainSpan: Atlas of the Developing Human Brain (Internet). Funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01, and 1RC2MH089929-01.
H.K.I. was supported by R01 MH107666-01.
Author information
Authors and Affiliations
Consortia
Contributions
L.M.H. designed the study and specific subanalyses, ran analyses, and wrote the manuscript. A.D. designed and ran analyses and contributed to the writing group. D.M.R. contributed to study and analytical design, and writing. G.H. contributed to analytical design and writing. W.W., H.T.N., and J.B. designed and ran specific analyses. A.F.P., V.M.R., T.D.A., K.G., M.F. all ran specific analyses. S.K.S. designed the study and analyses and contributed to the writing group. P.R. and R.K. designed the study and contributed data. E.D. designed the study, contributed data, and contributed to the writing group. E.R.G. designed specific analyses, and contributed to the writing group. S.P. designed the study. All three consortia (CMC, PGC-SCZ, iPSYCH-GEMS) contributed data. D.D., A.D.B., J.T.R.W., M.C.O’D., M.J.O. contributed data, advised on analyses, and contributed to the writing group. P. Sullivan advised on analyses and contributed to the writing group. B.D. designed the study, contributed data, advised on analyses, and contributed to the writing group. N.J.C. and H.K.I. designed the study, advised on analyses, and contributed to the writing group. P. Sklar and E.A.S. designed the study and specific analyses, ran analyses, and contributed to the writing group.
Corresponding author
Ethics declarations
Competing interests
E.D. has received research support from Roche during 2016–2018. T.W. has acted as advisor and lecturer to H. Lundbeck A/S. All other authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Supplementary Figures 1–12
Supplementary Table 1
Forward stepwise conditional analysis results.
Supplementary Table 2
MAGMA based pathway association results.
Supplementary Table 3
Mutant mouse lines lacking expression of SCZ-associated genes
Supplementary Table 4
Gene set membership of SCZ-associated genes, according to BRAINSPAN clusters
Rights and permissions
About this article
Cite this article
Huckins, L.M., Dobbyn, A., Ruderfer, D.M. et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51, 659–674 (2019). https://doi.org/10.1038/s41588-019-0364-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0364-4
This article is cited by
-
Prioritizing susceptibility genes for the prognosis of male-pattern baldness with transcriptome-wide association study
Human Genomics (2024)
-
Genomic insights into the comorbidity between type 2 diabetes and schizophrenia
Schizophrenia (2024)
-
Decreased CNNM2 expression in prefrontal cortex affects sensorimotor gating function, cognition, dendritic spine morphogenesis and risk of schizophrenia
Neuropsychopharmacology (2024)
-
Genetic imputation of kidney transcriptome, proteome and multi-omics illuminates new blood pressure and hypertension targets
Nature Communications (2024)
-
The genetic architecture of schizophrenia: review of large-scale genetic studies
Journal of Human Genetics (2023)