Abstract
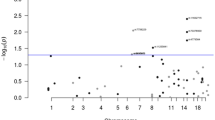
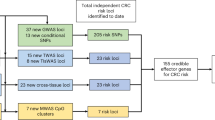
To further dissect the genetic architecture of colorectal cancer (CRC), we performed whole-genome sequencing of 1,439 cases and 720 controls, imputed discovered sequence variants and Haplotype Reference Consortium panel variants into genome-wide association study data, and tested for association in 34,869 cases and 29,051 controls. Findings were followed up in an additional 23,262 cases and 38,296 controls. We discovered a strongly protective 0.3% frequency variant signal at CHD1. In a combined meta-analysis of 125,478 individuals, we identified 40 new independent signals at P < 5 × 10−8, bringing the number of known independent signals for CRC to ~100. New signals implicate lower-frequency variants, Krüppel-like factors, Hedgehog signaling, Hippo-YAP signaling, long noncoding RNAs and somatic drivers, and support a role for immune function. Heritability analyses suggest that CRC risk is highly polygenic, and larger, more comprehensive studies enabling rare variant analysis will improve understanding of biology underlying this risk and influence personalized screening strategies and drug development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All whole-genome sequence data have been deposited in the database of Genotypes and Phenotypes (dbGaP), which is hosted by NCBI, under accession number phs001554.v1.p1. All custom Infinium OncoArray-500K array data for the studies in the stage 2 meta-analysis have been deposited at dbGaP under accession number phs001415.v1.p1. All Illumina HumanOmniExpressExome-8v1-2 array data for the studies in the stage 2 meta-analysis have been deposited at dbGaP under accession number phs001315.v1.p1. Genotype data for the studies included in the stage 1 meta-analysis have been deposited at dbGaP under accession number phs001078.v1.p1. The UK Biobank resource was accessed through application number 8614. CRC-relevant epigenome data were obtained from the NCBI Gene Expression Omnibus (GEO) database under accession number GSE77737.
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015).
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000).
Czene, K., Lichtenstein, P. & Hemminki, K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer 99, 260–266 (2002).
Sud, A., Kinnersley, B. & Houlston, R. S. Genome-wide association studies of cancer: current insights and future perspectives. Nat. Rev. Cancer 17, 692–704 (2017).
Tomlinson, I. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 39, 984–988 (2007).
Broderick, P. et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 39, 1315–1317 (2007).
Tomlinson, I. P. M. et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 40, 623–630 (2008).
Tenesa, A. et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 40, 631–637 (2008).
COGENT Study et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 40, 1426–1435 (2008).
Houlston, R. S. et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 42, 973–977 (2010).
Tomlinson, I. P. M. et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 7, e1002105 (2011).
Dunlop, M. G. et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet. 44, 770–776 (2012).
Peters, U. et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology 144, 799–807.e24 (2013).
Jia, W.-H. et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet. 45, 191–196 (2013).
Whiffin, N. et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 23, 4729–4737 (2014).
Wang, H. et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat. Commun. 5, 4613 (2014).
Zhang, B. et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 46, 533–542 (2014).
Schumacher, F. R. et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat. Commun. 6, 7138 (2015).
Al-Tassan, N. A. et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci. Rep. 5, 10442 (2015).
Orlando, G. et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum. Mol. Genet. 25, 2349–2359 (2016).
Zeng, C. et al. Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology 150, 1633–1645 (2016).
Schmit, S. L. et al. Novel common genetic susceptibility loci for colorectal cancer. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djy099 (2018).
Fuchsberger, C. et al. The genetic architecture of type 2 diabetes. Nature 536, 41–47 (2016).
1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Amos, C. I. et al. The Oncoarray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers. Prev. 26, 126–135 (2017).
Zhao, D. & DePinho, R. A. Synthetic essentiality: Targeting tumor suppressor deficiencies in cancer. Bioessays 39, (2017).
Zhao, D. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488 (2017).
Xiao, Y. et al. RGMb is a novel binding partner for PD-L2 and its engagement with PD-L2 promotes respiratory tolerance. J. Exp. Med. 211, 943–959 (2014).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Zhang, X. et al. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov. 8, 108–125 (2018).
Giannakis, M. et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 15, 857–865 (2016).
Dekker, R. J. et al. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood 107, 4354–4363 (2006).
Boon, R. A. et al. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler. Thromb. Vasc. Biol. 27, 532–539 (2007).
Chakroborty, D. et al. Dopamine stabilizes tumor blood vessels by up-regulating angiopoietin 1 expression in pericytes and Kruppel-like factor-2 expression in tumor endothelial cells. Proc. Natl Acad. Sci. USA 108, 20730–20735 (2011).
Lee, S.-J. et al. Regulation of hypoxia-inducible factor 1α (HIF-1α) by lysophosphatidic acid is dependent on interplay between p53 and Krüppel-like factor 5. J. Biol. Chem. 288, 25244–25253 (2013).
Zhang, H. et al. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Krüppel-like factor 5. J. Biol. Chem. 282, 15541–15549 (2007).
Ma, Z. et al. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget 8, 84153–84167 (2017).
Evangelista, M., Tian, H. & de Sauvage, F. J. The hedgehog signaling pathway in cancer. Clin. Cancer Res. 12, 5924–5928 (2006).
Gerling, M. et al. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat. Commun. 7, 12321 (2016).
Mille, F. et al. The Shh receptor Boc promotes progression of early medulloblastoma to advanced tumors. Dev. Cell. 31, 34–47 (2014).
Mathew, E. et al. Dosage-dependent regulation of pancreatic cancer growth and angiogenesis by hedgehog signaling. Cell Rep. 9, 484–494 (2014).
Zhao, B., Li, L., Lei, Q. & Guan, K.-L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 (2010).
Camargo, F. D. et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 (2007).
Ma, X., Zhang, H., Xue, X. & Shah, Y. M. Hypoxia-inducible factor 2α (HIF-2α) promotes colon cancer growth by potentiating Yes-associated protein 1 (YAP1) activity. J. Biol. Chem. 292, 17046–17056 (2017).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012).
Song, F. et al. Identification of a melanoma susceptibility locus and somatic mutation in TET2. Carcinogenesis 35, 2097–2101 (2014).
Eeles, R. A. et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 41, 1116–1121 (2009).
Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
Scott, L. J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Al Olama, A. A. et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 46, 1103–1109 (2014).
Timofeeva, M. N. et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet. 21, 4980–4995 (2012).
Shete, S. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 41, 899–904 (2009).
Bishop, D. T. et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 41, 920–925 (2009).
Sapkota, Y. et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 8, 15539 (2017).
Cannon-Albright, L. A. et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science 258, 1148–1152 (1992).
Hussussian, C. J. et al. Germline p16 mutations in familial melanoma. Nat. Genet. 8, 15–21 (1994).
Seoane, J. et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3, 400–408 (2001).
Jung, B., Staudacher, J. J. & Beauchamp, D. Transforming growth factor β superfamily signaling in development of colorectal cancer. Gastroenterology 152, 36–52 (2017).
Guda, K. et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc. Natl Acad. Sci. USA 106, 12921–12925 (2009).
Groden, J. et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66, 589–600 (1991).
Saharia, A. et al. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J. Biol. Chem. 285, 27057–27066 (2010).
Eeles, R. A. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 45, 385–391 (2013).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Paternoster, L. et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 47, 1449–1456 (2015).
Laken, S. J. et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat. Genet. 17, 79–83 (1997).
Niell, B. L., Long, J. C., Rennert, G. & Gruber, S. B. Genetic anthropology of the colorectal cancer-susceptibility allele APC I1307K: evidence of genetic drift within the Ashkenazim. Am. J. Hum. Genet. 73, 1250–1260 (2003).
Karami, S. et al. Telomere structure and maintenance gene variants and risk of five cancer types. Int. J. Cancer 139, 2655–2670 (2016).
Congrains, A., Kamide, K., Ohishi, M. & Rakugi, H. ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci. 14, 1278–1292 (2013).
Zhang, X. et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 48, 176–182 (2016).
Rheinbay, E. et al. Discovery and characterization of coding and non-coding driver mutations in more than 2,500 whole cancer genomes. Preprint at https://www.biorxiv.org/content/early/2017/12/23/237313 (2017).
Iotchkova, V. et al. GARFIELD - GWAS analysis of regulatory or functional information enrichment with LD correction. Preprint at https://www.biorxiv.org/content/early/2016/11/07/085738 (2016).
Segrè, A. V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Yang, J. et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 47, 1114–1120 (2015).
Bhatia, G. et al. Subtle stratification confounds estimates of heritability from rare variants. Preprint at https://www.biorxiv.org/content/early/2016/04/12/048181 (2016).
Zhong, H. & Prentice, R. L. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics 9, 621–634 (2008).
Cheetham, S. W., Gruhl, F., Mattick, J. S. & Dinger, M. E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 108, 2419–2425 (2013).
Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature 538, 161–164 (2016).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Jun, G., Wing, M. K., Abecasis, G. R. & Kang, H. M. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 25, 918–925 (2015).
Browning, B. L. & Yu, Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 85, 847–861 (2009).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Laurie, C. C. et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 34, 591–602 (2010).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at https://www.biorxiv.org/content/early/2017/07/20/166298 (2017).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Price, A. L. et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83, 132–135 (2008).
Weale, M. E. Quality control for genome-wide association studies. Methods Mol. Biol. 628, 341–372 (2010).
1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Delaneau, O., Howie, B., Cox, A. J., Zagury, J.-F. & Marchini, J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 93, 687–696 (2013).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Sun, J., Zheng, Y. & Hsu, L. A unified mixed-effects model for rare-variant association in sequencing studies. Genet. Epidemiol. 37, 334–344 (2013).
Moutsianas, L. et al. The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet. 11, e1005165 (2015).
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Cook, J. P., Mahajan, A. & Morris, A. P. Guidance for the utility of linear models in meta-analysis of genetic association studies of binary phenotypes. Eur. J. Hum. Genet. 25, 240–245 (2017).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361 (2013).
Wellcome Trust Case Control Consortium. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012).
Wakefield, J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 81, 208–227 (2007).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. Chapter 7, Unit7.20 (2013).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Quang, D., Chen, Y. & Xie, X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31, 761–763 (2015).
Ionita-Laza, I., McCallum, K., Xu, B. & Buxbaum, J. D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 48, 214–220 (2016).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Corradin, O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24, 1–13 (2014).
Pruitt, K. D. et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19, 1316–1323 (2009).
Harmston, N. et al. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8, 441 (2017).
Berlivet, S. et al. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9, e1004018 (2013).
Hu, Z. & Tee, W.-W. Enhancers and chromatin structures: regulatory hubs in gene expression and diseases. Biosci. Rep. 37, BSR20160183 (2017).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Witte, J. S., Visscher, P. M. & Wray, N. R. The contribution of genetic variants to disease depends on the ruler. Nat. Rev. Genet. 15, 765–776 (2014).
Cox, A. et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 39, 352–358 (2007).
Johns, L. E. & Houlston, R. S. A systematic review and meta-analysis of familial colorectal cancer risk. Am. J. Gastroenterol. 96, 2992–3003 (2001).
Hsu, L. et al. A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology 148, 1330–1339.e14 (2015).
Jeon, J. et al. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology 154, 2152–2164.e19 (2018).
Acknowledgements
A full list of acknowledgements appears in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
J.R.H., S.A.B., T.A.H., H.M.K., D.V.C., M.W., F.R.S., J.D.S., D.A., M.H.A., K.A., C.A.-C., V.A., C.B., J.A.B., S.I.B., S.B., D.T.B., J.B., H.Boeing, H.Brenner, S.Brezina, S.Buch, D.D.B., A.B.-H., K.B., B.J.C., P.T.C., S.C.-B., A.T.C., J.C.-C., S.J.C., M.-D.C., S.H.C., A.J.C., K.C., A.d.l.C., D.F.E., S.G.E., F.E., D.R.E., E.J.M.F., J.C.F., D.F., S.G., G.G.G., E.G., P.J.G., J.S.G., A.G., M.J.G., R.W.H., J.H., H.H., R.B.H., P.H., M.H., J.L.H., W.-Y.H., T.J.H., D.J.H., R.J., E.J.J., M.A.J., T.O.K., T.J.K., H.R.K., L.N.K., C.K., S.K., S.-S.K., L.L.M., S.C.L., C.I.L., L.L., A.L., N.M.L., S.M., S.D.M., V.M., G.M., M.M., R.L.M., L.M., R.M., A.N., P.A.N., K.O., N.C.O.-M., B.P., P.S.P., R.P., V.P., P.D.P.P., E.A.P., R.L.P., G.R., H.S.R., E.R., M.R.-B., C.S., R.E.S., D.S., M.-H.S., S.S., M.L.S., C.M.T., S.N.T., A.T., C.M.U., F.J.B.v.D., B.V.G., H.v.K., J.V., K.V., P.V., L.V., V.V., E.W., C.R.W., A.W., M.O.W., A.H.W., B.W.Z., W.Z., P.C.S., J.D.P., M.C.B., G.C., V.M., G.R.A., D.A.N., S.B.G., L.H. and U.P. conceived and designed the experiments. T.A.H., M.W., J.D.S., K.F.D., D.D., R.I., E.K., H.L., C.E.M., E.P., J.R., T.S., S.S.T., D.J.V.D.B., M.C.B. and D.A.N. performed the experiments. J.R.H., H.M.K., S.C., S.L.S., D.V.C., C.Q., J.J., C.K.E., P.G., F.R.S., D.M.L., S.C.N., N.A.S.-A., C.A.L., M.L., T.L.L., Y.-R.S., A.K., G.R.A. and L.H. performed statistical analysis. J.R.H., S.A.B., T.A.H., H.M.K., S.C., S.L.S., D.V.C., C.Q., J.J., C.K.E., P.G., M.W., F.R.S., D.M.L., S.C.N., N.A.S.-A., B.L.B., C.S.C., C.M.C., K.R.C., J.G., W.-L.H., C.A.L., S.M.L., M.L., Y.L., T.L.L., M.S., Y.-R.S., A.K., G.R.A., L.H. and U.P. analyzed the data. H.M.K., C.K.E., D.A., M.H.A., K.A., C.A.-C., V.A., C.B., J.A.B., S.I.B., S.B., D.T.B., J.B., H.Boeing, H.Brenner, S.Brezina, S.Buch, D.D.B., A.B.-H., K.B., B.J.C., P.T.C., S.C.-B., A.T.C., J.C.-C., S.J.C., M.-D.C., S.H.C., A.J.C., K.C., A.d.l.C., D.F.E., S.G.E., F.E., D.R.E., E.J.M.F., J.C.F., R.F., L.M.F., D.F., M.G., S.G., W.J.G., G.G.G., P.J.G., W.M.G., J.S.G., A.G., M.J.G., R.W.H., J.H., H.H., S.H., R.B.H., P.H., M.H., J.L.H., W.-Y.H., T.J.H., D.J.H., G.I.-S., G.E.I., R.J., E.J.J., M.A.J., A.D.J., C.E.J., T.O.K., T.J.K., H.R.K., L.N.K., C.K., T.K., S.K., S.-S.K., S.C.L., L.L.M., S.C.L., F.L., C.I.L., L.L., W.L., A.L., N.M.L., S.M., S.D.M., V.M., G.M., M.M., R.L.M., L.M., N.M., R.M., A.N., P.A.N., K.O., S.O, N.C.O.-M., B.P., P.S.P., R.P., V.P., P.D.P.P., M.P., E.A.P., R.L.P., L.R., G.R., H.S.R., E.R., M.R.-B., L.C.S., C.S., R.E.S., M.S., M.-H.S., K.S., S.S., M.L.S., M.C.S., Z.K.S., C.S., C.M.T., S.N.T., D.C.T., A.E.T., A.T., C.M.U., F.J.B.v.D., B.V.G., H.v.K., J.V., K.V., P.V., L.V., V.V., K.W., S.J.W., E.W., A.K.W., C.R.W., A.W., M.O.W., A.H.W., S.H.Z., B.W.Z., Q.Z., W.Z., P.C.S., J.D.P., M.C.B., A.K., G.C., V.M., G.R.A., S.B.G. and U.P. contributed reagents, materials and analysis tools. J.R.H., S.A.B., T.A.H., J.J., L.H. and U.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
G.R.A. has received compensation from 23andMe and Helix. He is currently an employee of Regeneron Pharmaceuticals. H.H. performs collaborative research with Ambry Genetics, InVitae Genetics, and Myriad Genetic Laboratories, is on the scientific advisory board for InVitae Genetics and Genome Medical, and has stock in Genome Medical. R.P. has participated in collaborative funded research with Myriad Genetics Laboratories and Invitae Genetics but has no financial competitive interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note
Supplementary Table 1
Characteristics of studies and study participants contributing to the whole-genome sequencing analysis and GWAS meta-analysis
Supplementary Table 2
Association results broken down by sample set together with imputation qualities, and heterogeneity statistics for new loci reported in Table 1
Supplementary Table 3
Colorectal cancer risk signals previously associated at genome-wide significance
Supplementary Table 4
Conditional association results broken down by sample set together with imputation qualities, and heterogeneity statistics for new conditionally independent association signals reported in Table 2
Supplementary Table 5
Known and newly identified CRC risk loci with multiple conditionally independent association signals that reach a significance threshold of P < 1 × 10–5 in the combined meta-analysis of up to 125,478 individuals
Supplementary Table 6
Reported associations of colorectal cancer risk variants with (non-colorectal cancer) diseases and traits in the NHGRI-EBI GWAS catalog
Supplementary Table 7
Summary of 99% credible sets for the 40 new association signals for colorectal cancer risk
Supplementary Table 8
CRC relevant annotations, bioinformatic follow-up of newly identified loci, and bioinformatic follow-up of secondary signals
Supplementary Table 9
Enrichment of CRC risk associations in 1,005 genomic annotations from the ENCODE, Roadmap Epigenomics and GENCODE projects at the 1 × 10–5 and 1 × 10–8 significance thresholds
Supplementary Table 10
MAGENTA pathway enrichment results
Supplementary Table 11
Risk allele frequencies (RAFs) across populations for the 95 variants used in the polygenic risk score analyses
Supplementary Table 12
Covariates included in the association analysis
Supplementary Table 13
CRC relevant regulatory genomic datasets
Supplementary Table 14
Results from ATAC-QC
Supplementary Table 15
Colorectal cancer risk variants and effect size estimates used in the familial risk explained and genetic risk score analyses
Rights and permissions
About this article
Cite this article
Huyghe, J.R., Bien, S.A., Harrison, T.A. et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet 51, 76–87 (2019). https://doi.org/10.1038/s41588-018-0286-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0286-6
This article is cited by
-
Genome-wide polygenic risk scores for colorectal cancer have implications for risk-based screening
British Journal of Cancer (2024)
-
Polygenic risk scores, radiation treatment exposures and subsequent cancer risk in childhood cancer survivors
Nature Medicine (2024)
-
Prioritization of risk genes in colorectal cancer by integrative analysis of multi-omics data and gene networks
Science China Life Sciences (2024)
-
Cellular senescence gene TACC3 associated with colorectal cancer risk via genetic and DNA methylated alteration
Archives of Toxicology (2024)
-
Allometric versus traditional body-shape indices and risk of colorectal cancer: a Mendelian randomization analysis
International Journal of Obesity (2024)