Abstract
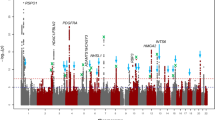
Refractive errors, including myopia, are the most frequent eye disorders worldwide and an increasingly common cause of blindness. This genome-wide association meta-analysis in 160,420 participants and replication in 95,505 participants increased the number of established independent signals from 37 to 161 and showed high genetic correlation between Europeans and Asians (>0.78). Expression experiments and comprehensive in silico analyses identified retinal cell physiology and light processing as prominent mechanisms, and also identified functional contributions to refractive-error development in all cell types of the neurosensory retina, retinal pigment epithelium, vascular endothelium and extracellular matrix. Newly identified genes implicate novel mechanisms such as rod-and-cone bipolar synaptic neurotransmission, anterior-segment morphology and angiogenesis. Thirty-one loci resided in or near regions transcribing small RNAs, thus suggesting a role for post-transcriptional regulation. Our results support the notion that refractive errors are caused by a light-dependent retina-to-sclera signaling cascade and delineate potential pathobiological molecular drivers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pan, C. W., Ramamurthy, D. & Saw, S. M. Worldwide prevalence and risk factors for myopia. Ophthalmic Physiol. Opt. 32, 3–16 (2012).
Morgan, I. G. What public policies should be developed to deal with the epidemic of myopia? Optom. Vis. Sci. 93, 1058–1060 (2016).
Morgan, I. & Rose, K. How genetic is school myopia? Prog. Retin. Eye Res. 24, 1–38 (2005).
Morgan, I. G., Ohno-Matsui, K. & Saw, S. M. Myopia. Lancet 379, 1739–1748 (2012).
Williams, K. M. et al. Increasing prevalence of myopia in Europe and the impact of education. Ophthalmology 122, 1489–1497 (2015).
Williams, K. M. et al. Prevalence of refractive error in Europe: the European Eye Epidemiology (E(3)) Consortium. Eur. J. Epidemiol. 30, 305–315 (2015).
Vongphanit, J., Mitchell, P. & Wang, J. J. Prevalence and progression of myopic retinopathy in an older population. Ophthalmology 109, 704–711 (2002).
Seet, B. et al. Myopia in Singapore: taking a public health approach. Br. J. Ophthalmol. 85, 521–526 (2001).
Smith, T. S., Frick, K. D., Holden, B. A., Fricke, T. R. & Naidoo, K. S. Potential lost productivity resulting from the global burden of uncorrected refractive error. Bull. World Health Organ. 87, 431–437 (2009).
Verhoeven, V. J. et al. Visual consequences of refractive errors in the general population. Ophthalmology 122, 101–109 (2015).
Tideman, J. W. et al. Association of axial length with risk of uncorrectable visual impairment for Europeans with myopia. JAMA Ophthalmol. 134, 1355–1363 (2016).
Flitcroft, D. I. The complex interactions of retinal, optical and environmental factors in myopia aetiology. Prog. Retin. Eye Res. 31, 622–660 (2012).
Nakanishi, H. et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 5, e1000660 (2009).
Lam, C. Y. et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest. Ophthalmol. Vis. Sci. 49, 3768–3778 (2008).
Stambolian, D. et al. Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum. Mol. Genet. 22, 2754–2764 (2013).
Fan, Q. et al. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 8, e1002753 (2012).
Fan, Q. et al. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat. Commun. 7, 11008 (2016).
Cheng, C. Y. et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am. J. Hum. Genet. 93, 264–277 (2013).
Shi, Y. et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 7, e1002084 (2011).
Shi, Y. et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am. J. Hum. Genet. 88, 805–813 (2011).
Li, Y. J. et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology 118, 368–375 (2011).
Li, Z. et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum. Mol. Genet. 20, 2861–2868 (2011).
Liu, J. & Zhang, H. X. Polymorphism in the 11q24.1 genomic region is associated with myopia: a comprehensive genetic study in Chinese and Japanese populations. Mol. Vis. 20, 352–358 (2014).
Tran-Viet, K. N. et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am. J. Hum. Genet. 92, 820–826 (2013).
Aldahmesh, M. A. et al. Mutations in LRPAP1 are associated with severe myopia in humans. Am. J. Hum. Genet. 93, 313–320 (2013).
Verhoeven, V. J. et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 45, 314–318 (2013).
Kiefer, A. K. et al. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 9, e1003299 (2013).
Wojciechowski, R. & Hysi, P. G. Focusing in on the complex genetics of myopia. PLoS Genet. 9, e1003442 (2013).
1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Plotnikov, D., Guggenheim, J. & The UK Biobank Eye and Vision Consortium. Is a large eye size a risk factor for myopia? A Mendelian randomization study. https://www.biorxiv.org/content/early/2017/12/29/240283/ (2017).
Hsu, F. et al. The UCSC Known Genes. Bioinformatics 22, 1036–1046 (2006).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Kelly, M. P. Does phosphodiesterase 11A (PDE11A) hold promise as a future therapeutic target? Curr. Pharm. Des. 21, 389–416 (2015).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Mathe, E. et al. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 34, 1317–1325 (2006).
Tavtigian, S. V. et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 43, 295–305 (2006).
Bakshi, A. et al. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 6, 32894 (2016).
Ferreira, M. A. et al. Gene-based analysis of regulatory variants identifies 4 putative novel asthma risk genes related to nucleotide synthesis and signaling. J. Allergy Clin. Immunol. 139, 1148–1157 (2017).
Pickrell, J. K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94, 559–573 (2014).
Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Verhoeven, V. J. et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum. Genet. 131, 1467–1480 (2012).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Brown, B. C., Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Pers, T. H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
Fritsche, L. G. et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 48, 134–143 (2016).
Ritchey, E. R. et al. Vision-guided ocular growth in a mutant chicken model with diminished visual acuity. Exp. Eye Res. 102, 59–69 (2012).
Vincent, A. et al. Biallelic mutations in GNB3 cause a unique form of autosomal-recessive congenital stationary night blindness. Am. J. Hum. Genet. 98, 1011–1019 (2016).
Blake, J. A. et al. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 45, D723–D729 (2017).
Nikonov, S. S. et al. Cones respond to light in the absence of transducin β subunit. J. Neurosci. 33, 5182–5194 (2013).
Stone, E. M. et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 22, 199–202 (1999).
Mackay, D. S., Bennett, T. M. & Shiels, A. Exome sequencing identifies a missense variant in EFEMP1 co-segregating in a family with autosomal dominant primary open-angle glaucoma. PLoS One 10, e0132529 (2015).
Springelkamp, H. et al. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum. Mol. Genet. 24, 2689–2699 (2015).
Haeseleer, F. et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat. Neurosci. 7, 1079–1087 (2004).
Littink, K. W. et al. A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Invest. Ophthalmol. Vis. Sci. 50, 2344–2350 (2009).
Grimes, W. N., Li, W., Chávez, A. E. & Diamond, J. S. BK channels modulate pre- and postsynaptic signaling at reciprocal synapses in retina. Nat. Neurosci. 12, 585–592 (2009).
Keckeis, S., Reichhart, N., Roubeix, C. & Strauß, O. Anoctamin2 (TMEM16B) forms the Ca2+-activated Cl– channel in the retinal pigment epithelium. Exp. Eye Res. 154, 139–150 (2017).
Prasanna, G., Narayan, S., Krishnamoorthy, R. R. & Yorio, T. Eyeing endothelins: a cellular perspective. Mol. Cell. Biochem. 253, 71–88 (2003).
Yamashita, T. et al. Essential and synergistic roles of RP1 and RP1L1 in rod photoreceptor axoneme and retinitis pigmentosa. J. Neurosci. 29, 9748–9760 (2009).
Davidson, A. E. et al. RP1L1 variants are associated with a spectrum of inherited retinal diseases including retinitis pigmentosa and occult macular dystrophy. Hum. Mutat. 34, 506–514 (2013).
Hawthorne, F. et al. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Invest. Ophthalmol. Vis. Sci. 54, 2076–2086 (2013).
Feldkaemper, M. & Schaeffel, F. An updated view on the role of dopamine in myopia. Exp. Eye Res. 114, 106–119 (2013).
Paul, M. L., Graybiel, A. M., David, J. C. & Robertson, H. A. D1-like and D2-like dopamine receptors synergistically activate rotation and c-fos expression in the dopamine-depleted striatum in a rat model of Parkinson’s disease. J. Neurosci. 12, 3729–3742 (1992).
Stone, R. A., Lin, T., Laties, A. M. & Iuvone, P. M. Retinal dopamine and form-deprivation myopia. Proc. Natl. Acad. Sci. USA 86, 704–706 (1989).
Gardner, M., Bertranpetit, J. & Comas, D. Worldwide genetic variation in dopamine and serotonin pathway genes: implications for association studies. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147B, 1070–1075 (2008).
D’Souza, U. M. & Craig, I. W. Functional polymorphisms in dopamine and serotonin pathway genes. Hum. Mutat. 27, 1–13 (2006).
Beaulieu, J. M. & Gainetdinov, R. R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 63, 182–217 (2011).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Holden, B. A. et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology 123, 1036–1042 (2016).
Cardon, L. R. & Palmer, L. J. Population stratification and spurious allelic association. Lancet 361, 598–604 (2003).
Chua, S. Y. et al. Age of onset of myopia predicts risk of high myopia in later childhood in myopic Singapore children. Ophthalmic Physiol. Opt. 36, 388–394 (2016).
Williams, K. M. et al. Age of myopia onset in a British population-based twin cohort. Ophthalmic Physiol. Opt. 33, 339–345 (2013).
Dolgin, E. The myopia boom. Nature 519, 276–278 (2015).
Connaughton, V. Glutamate and glutamate receptors in the vertebrate retina. In: H. Kolb et al. eds. Webvision: The Organization of the Retina and Visual System (Webvision, Salt Lake City, UT, USA, 1995).
Hung, G. K., Mahadas, K. & Mohammad, F. Eye growth and myopia development: unifying theory and Matlab model. Comput. Biol. Med. 70, 106–118 (2016).
Norton, T. T. What do animal studies tell us about the mechanism of myopia-protection by light? Optom. Vis. Sci. 93, 1049–1051 (2016).
Weiss, S. & Schaeffel, F. Diurnal growth rhythms in the chicken eye: relation to myopia development and retinal dopamine levels. J. Comp. Physiol. A 172, 263–270 (1993).
Stone, R. A., Lin, T., Iuvone, P. M. & Laties, A. M. Postnatal control of ocular growth: dopaminergic mechanisms. Ciba Found. Symp. 155, 45–62 (1990).
Morgan, I. G. The biological basis of myopic refractive error. Clin. Exp. Optom. 86, 276–288 (2003).
Li, X. X., Schaeffel, F., Kohler, K. & Zrenner, E. Dose-dependent effects of 6-hydroxy dopamine on deprivation myopia, electroretinograms, and dopaminergic amacrine cells in chickens. Vis. Neurosci. 9, 483–492 (1992).
Iuvone, P. M., Tigges, M., Stone, R. A., Lambert, S. & Laties, A. M. Effects of apomorphine, a dopamine receptor agonist, on ocular refraction and axial elongation in a primate model of myopia. Invest. Ophthalmol. Vis. Sci. 32, 1674–1677 (1991).
Ashby, R., McCarthy, C. S., Maleszka, R., Megaw, P. & Morgan, I. G. A muscarinic cholinergic antagonist and a dopamine agonist rapidly increase ZENK mRNA expression in the form-deprived chicken retina. Exp. Eye Res. 85, 15–22 (2007).
Ashby, R. Animal studies and the mechanism of myopia-protection by light? Optom. Vis. Sci. 93, 1052–1054 (2016).
Rymer, J. & Wildsoet, C. F. The role of the retinal pigment epithelium in eye growth regulation and myopia: a review. Vis. Neurosci. 22, 251–261 (2005).
Chen, S. et al. Bright light suppresses form-deprivation myopia development with activation of dopamine D1 receptor signaling in the ON pathway in retina. Invest. Ophthalmol. Vis. Sci. 58, 2306–2316 (2017).
Chen, P. S. et al. Effects of C825T polymorphism of the GNB3 gene on availability of dopamine transporter in healthy volunteers: a SPECT study. Neuroimage 56, 1526–1530 (2011).
Scott, M. S. & Ono, M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie 93, 1987–1992 (2011).
McFadden, S. A. Understanding and treating myopia: what more we need to know and future research priorities. Optom. Vis. Sci. 93, 1061–1063 (2016).
Smith, E. L. III, Hung, L. F. & Arumugam, B. Visual regulation of refractive development: insights from animal studies. Eye (Lond.) 28, 180–188 (2014).
Zhang, Y. & Wildsoet, C. F. RPE and choroid mechanisms underlying ocular growth and myopia. Prog. Mol. Biol. Transl. Sci. 134, 221–240 (2015).
Harper, A. R. & Summers, J. A. The dynamic sclera: extracellular matrix remodeling in normal ocular growth and myopia development. Exp. Eye Res. 133, 100–111 (2015).
Summers, J. A. The choroid as a sclera growth regulator. Exp. Eye Res. 114, 120–127 (2013).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Chen, W. M. & Abecasis, G. R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81, 913–926 (2007).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Zaykin, D. V. Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J. Evol. Biol. 24, 1836–1841 (2011).
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Yang, H. & Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10, 1556–1566 (2015).
Cooper, G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005).
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Consortium, G. T., GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Bauer-Mehren, A., Rautschka, M., Sanz, F. & Furlong, L. I. DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics 26, 2924–2926 (2010).
Günther, S. et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 36, D919–D922 (2008).
Kuhn, M. et al. STITCH 4: integration of protein-chemical interactions with user data. Nucleic Acids Res. 42, D401–D407 (2014).
Wishart, D. S. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006).
Whirl-Carrillo, M. et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417 (2012).
Acknowledgements
We gratefully thank all study participants, their relatives and the staff at the recruitment centers for their invaluable contributions. We thank all contributors to the CREAM Consortium, 23andMe and UKEV for their generosity in sharing data and help in the production of this publication. Funding for this particular GWAS mega-analysis was provided by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant 648268), the Netherlands Organisation for Scientific Research (NWO, grant 91815655) and the National Eye Institute (grant R01EY020483). Funding agencies that facilitated the execution of the individual studies are acknowledged in the Supplementary Note.
The Consortium for Refractive Error and Myopia (CREAM Consortium):
Tin Aung82,83, Amutha B. Veluchamy82,84, Kathryn P. Burdon58, Harry Campbell36, Li Jia Chen85, Peng Chen83, Wei Chen86, Emily Chew45, Margaret M. Deangelis87, Xiaohu Ding88, Angela Döring66, David M. Evans89,90, Sheng Feng91, Brian Fleck92, Rhys D. Fogarty58, Jeremy R. Fondran43, Maurizio Fossarello93, Xiaobo Guo88,94, Annet E. G. Haarman1,2, Mingguang He23,88, Laura D. Howe90,95, Sarayut Janmahasatian43, Vishal Jhanji85, Mika Kähönen96, Jaakko Kaprio20,97, John P. Kemp90, Kay-Tee Khaw11, Chiea-Chuen Khor29,83,87,98, Eva Krapohl99, Jean-François Korobelnik100,101, Kris Lee9, Shi-Ming Li22, Yi Lu56, Robert N. Luben11, Kari-Matti Mäkelä49, George McMahon90, Akira Meguro102, Evelin Mihailov18, Masahiro Miyake16, Nobuhisa Mizuki102, Margaux Morrison87, Vinay Nangia103, Konrad Oexle104, Songhomitra Panda-Jonas103, Chi Pui Pang85, Mario Pirastu105, Robert Plomin99, Taina Rantanen77, Maria Schache23, Ilkka Seppälä49, George D. Smith90, Beate St Pourcain90,106, Pancy O. Tam85, J. Willem L. Tideman1,2, Nicholas J. Timpson90, Simona Vaccargiu105, Zoran Vatavuk35, Jie Jin Wang23,24, Ningli Wang22, Nick J. Wareham107, Alan F. Wright33, Liang Xu22, Maurice K. H. Yap108, Seyhan Yazar74, Shea Ping Yip109, Nagahisa Yoshimura16, Alvin L. Young9, Jing Hua Zhao107 and Xiangtian Zhou86 UK Biobank Eye and Vision Consortium: Tariq M. Aslam110, Sarah A. Barman111, Jenny H. Barrett112, Paul N. Bishop110, Peter Blows12, Catey Bunce113, Roxana O. Carare114, Usha Chakravarthy115, Michelle Chan12, Sharon Chua12, David Crabb116, Alexander Day12, Parul Desai12, Bal Dhillon117, Andrew D. Dick118, Cathy A. Egan12, Sarah Ennis114, Marcus Fruttiger12, John Gallacher119, David F. Garway-Heath12, Jane Gibson114, Dan M. Gore12, Alison Hardcastle12, Simon P. Harding120, Ruth E. Hogg121, Pearse A. Keane12, Peng Tee Khaw12, Gerassimos Lascaratos12, Andrew Lotery122, Phil J. Luthert12, Tom J. MacGillivray123, Sarah L. Mackie124, Keith R. Martin125, Michelle McGaughey126, Bernadette McGuinness126, Gareth J. McKay126, Martin McKibbin127, Danny Mitry12, Tony Moore12, James E. Morgan26, Zaynah A. Muthy12, Eoin O’Sullivan128, Chris Owen129, Praveen J. Patel12, Euan N. Paterson126, Tunde Peto115, Axel Petzold130, Alicja R. Rudnicka129, Jay E. Self122,131, Sobha Sivaprasad12, David H. W. Steel132, Irene M. Stratton133, Nicholas Strouthidis12,Cathie L. M. Sudlow134, Caroline Thaung12, Dhanes Thomas12, Emanuele Trucco135, Adnan Tufail12, Stephen A. Vernon136, Ananth C. Viswanathan12, Jayne V. Woodside126, Max Yates137, Jennifer L. Y. Yip11 and Yalin Zheng120 23andMe Research Team: Michelle Agee7, Babak Alipanahi7, Adam Auton7, Robert K. Bell7, Katarzyna Bryc7, Sarah L. Elson7, Pierre Fontanillas7, David A. Hinds7, Jennifer C. McCreight7, Karen E. Huber7, Aaron Kleinman7, Nadia K. Litterman7, Matthew H. McIntyre7, Joanna L. Mountain7, Elizabeth S. Noblin7, Carrie A. M. Northover7, Steven J. Pitts7, J. Fah Sathirapongsasuti7, Olga V. Sazonova7, Janie F. Shelton7, Suyash Shringarpure7, Chao Tian7, Vladimir Vacic7 and Catherine H. Wilson7
Author information
Authors and Affiliations
Consortia
Contributions
M.S.T., V.J.M.V., S.M., J.A.G., A.I.I., R.W., P.G.H., A.I.I. and E.M.v.L. performed the analyses. C.C.W.K., V.J.M.V., M.S.T., R.W., J.A.G. and S.M. drafted the manuscript, and C.J.H., P.G.H., A.P.K., C.M.v.D., D.S., E.M.v.L., J.E.B.-W., J.Y.T., N.A.F., Q.F., S.-M.S. and V.V. critically reviewed the manuscript. A.N., A.P.K., A.T., C.B., C. Gieger, C.L.S., C.-Y.C., G. Biino, G.C.-P., I.R., J.E.B.W., J.E.H., J. S. Ried, J.W., J.X., K.M.W., K.Y., P.M.C., S.M.H., M.S.T., N.A.F., N.E., P.C., P. Gharahkhani, P.K.J., Q.F., R. Höhn, R.L.S., R.P.I., R.W., T.H., T.-H.S.-A., T.Z., V.V., W.-Y.S., W.Z., X.L.S., Y.C.T., Y.S. and Y.Y.T. performed data analysis for the individual studies; A.D.P., A.G.U., A.T., A.W.H., B.E.K.K., C.C.W.K., C.D., C. Grazal, C.H., C.J.H., C.W., C.-Y.C., D.A.M., F.R., G. Bencic, H.M.-H., J.A.G., J.B.J., J.E.B.-W., J.E.C., J.F.W., J.H.L., J.R.V., J. S. Rahi, J. S. Ried, J.Y.T., K.Y., M.A.M.-S., N.G.M., N.P., O. Polašek, O. Pärssinen, O.T.R., P. Gupta, P.J.F., P.M., P.N.B., R.K., S.K.I., S.-M.S., T.L., T.M., W.Z., Y.C.T. and Y.X.W. contributed to data assembly. A.A.B.B., A.W., C. Grazal, D.S., K.N.W., S.W.T. and T.L.Y. performed expression experiments, and M.S.T., A.A.B.B., P.J.v.d.S. and R. Hask performed in silico pathway analyses. C.C.W.K. and C.J.H. conceived and designed the outline of the current report, and supervised conduction of experiments and analyses jointly with A.M., A.H., A.W.H., C.D., C.H., C.J.H., C.M.v.D., C.W., C.-Y.C., D.A.M., D.S., E.-S.T., F.M., G. Biino, I.R., J.A.G., J.B.J., J.E.B.-W., J.E.C., J.F.W., J.H.L., J.R.V., J.Y.T., N.A., N.A.F., N.P., O. Pärssinen, O.T.R., P.J.F., P.N.B., S.K.I., S.-M.S., T.L., T.Y.W., T.L.Y., V.V., Y.X.W. and Y.Y.T. M.P.C. analyzed the data and performed statistical analyses. The 23andMe research team, CREAM and the UK Biobank Eye and Vision Consortium contributed reagents/materials/analysis tools and performed statistical analyses.
Corresponding author
Ethics declarations
Competing interests
N.A.F., N.E., J.Y.T. and the 23andMe Research Team are current or former employees of 23andMe, Inc., and hold stock or stock options in 23andMe. J.B.J. is a patent holder with Biocompatibles UK Ltd. (Franham, Surrey, UK) (Title: Treatment of eye diseases using encapsulated cells encoding and secreting neuroprotective factor and /or anti-angiogenic factor; international patent no. 20120263794) and is included in a patent application with University of Heidelberg (Heidelberg, Germany) (Title: Agents for use in the therapeutic or prophylactic treatment of myopia or hyperopia; European patent no. 3 070 101). The other authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note and Supplementary Tables 1, 5–9, 12 and 17
Supplementary Table 2
Stage 1 - 3 Meta-Analyses and Conditional Analysis
Supplementary Table 3
Stage 3 Index Variants of FE and RE analyses
Supplementary Table 4
HWE p-value per cohort TopSNPs Stage 3
Supplementary Table 10
DEPICT analysis - gene-set enrichment
Supplementary Table 11
Ranking of associated genes according to biological plausibility
Supplementary Table 13
Genes with Human Ocular Phenotypes
Supplementary Table 14
Genes with Mice Ocular Phenotypes
Supplementary Table 15
Expression of Candidate Genes in Ocular Tissues
Supplementary Table 16
Genetic variants harboring known drug targets
Supplementary Data 1
Locus Zoom Plots
Supplementary Data 2
Forest Plots Stage 3 and Conditional Loci
Supplementary Data 3
Summary Statistics Stage 3 Meta-analysis
Appendices
The CREAM Consortium
Tin Aung82,83, Amutha B. Veluchamy82, 84, Kathryn P. Burdon58, Harry Campbell36, Li Jia Chen85, Peng Chen83, Wei Chen86, Emily Chew45, Margaret M. Deangelis87, Xiaohu Ding88, Angela Döring66, David M. Evans89, 90, Sheng Feng91, Brian Fleck92, Rhys D. Fogarty58, Jeremy R. Fondran43, Maurizio Fossarello93, Xiaobo Guo88, 94, Annet E. G. Haarman1, 2, Mingguang He23, 88, Laura D. Howe90, 95, Sarayut Janmahasatian43, Vishal Jhanji85, Mika Kähönen96, Jaakko Kaprio20, 97, John P. Kemp90, Kay-Tee Khaw11, Chiea-Chuen Khor29, 83, 87, 98, Eva Krapohl99, Jean-François Korobelnik100, 101, Kris Lee9, Shi-Ming Li22, Yi Lu56, Robert N. Luben11, Kari-Matti Mäkelä49, George McMahon90, Akira Meguro102, Evelin Mihailov18, Masahiro Miyake16, Nobuhisa Mizuki102, Margaux Morrison87, Vinay Nangia103, Konrad Oexle104, Songhomitra Panda-Jonas103, Chi Pui Pang85, Mario Pirastu105, Robert Plomin99, Taina Rantanen77, Maria Schache23, Ilkka Seppälä49, George D. Smith90, Beate St Pourcain90, 106, Pancy O. Tam85, J. Willem L. Tideman1, 2, Nicholas J. Timpson90, Simona Vaccargiu105, Zoran Vatavuk35, Jie Jin Wang23, 24, Ningli Wang22, Nick J. Wareham107, Alan F. Wright33, Liang Xu22, Maurice K. H. Yap108, Seyhan Yazar74, Shea Ping Yip109, Nagahisa Yoshimura16, Alvin L. Young9, Jing Hua Zhao107 and Xiangtian Zhou86
23andMe Research Team
Michelle Agee7, Babak Alipanahi7, Adam Auton7, Robert K. Bell7, Katarzyna Bryc7, Sarah L. Elson7, Pierre Fontanillas7, David A. Hinds7, Jennifer C. McCreight7, Karen E. Huber7, Aaron Kleinman7, Nadia K. Litterman7, Matthew H. McIntyre7, Joanna L. Mountain7, Elizabeth S. Noblin7, Carrie A. M. Northover7, Steven J. Pitts7, J. Fah Sathirapongsasuti7, Olga V. Sazonova7, Janie F. Shelton7, Suyash Shringarpure7, Chao Tian7, Vladimir Vacic7 and Catherine H. Wilson7
UK Biobank Eye and Vision Consortium
Tariq M. Aslam110, Sarah A. Barman111, Jenny H. Barrett112, Paul N. Bishop110, Peter Blows12, Catey Bunce113, Roxana O. Carare114, Usha Chakravarthy115, Michelle Chan12, Sharon Chua12, David Crabb116, Alexander Day12, Parul Desai12, Bal Dhillon117, Andrew D. Dick118, Cathy A. Egan12, Sarah Ennis114, Marcus Fruttiger12, John Gallacher119, David F. Garway-Heath12, Jane Gibson114, Dan M. Gore12, Alison Hardcastle12, Simon P. Harding120, Ruth E. Hogg121, Pearse A. Keane12, Peng Tee Khaw12, Gerassimos Lascaratos12, Andrew Lotery122, Phil J. Luthert12, Tom J. MacGillivray123, Sarah L. Mackie124, Keith R. Martin125, Michelle McGaughey126, Bernadette McGuinness126, Gareth J. McKay126, Martin McKibbin127, Danny Mitry12, Tony Moore12, James E. Morgan26, Zaynah A. Muthy12, Eoin O’Sullivan128, Chris Owen129, Praveen J. Patel12, Euan N. Paterson126, Tunde Peto115, Axel Petzold130, Alicja R. Rudnicka129, Jay E. Self122,131, Sobha Sivaprasad12, David H. W. Steel132, Irene M. Stratton133, Nicholas Strouthidis12, Cathie L. M. Sudlow134, Caroline Thaung12, Dhanes Thomas12, Emanuele Trucco135, Adnan Tufail12, Stephen A. Vernon136, Ananth C. Viswanathan12, Jayne V. Woodside126, Max Yates137, Jennifer L. Y. Yip11 and Yalin Zheng120
82Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Singapore. 83Department of Ophthalmology, National University HealthSystems, National University of Singapore, Singapore, Singapore. 84Duke-NUS Medical School, Singapore, Singapore, Singapore. 85Department ofOphthalmology and Visual Sciences, Chinese University of Hong Kong, Hong Kong Eye Hospital, Kowloon, Hong Kong. 86School of Ophthalmologyand Optometry, Eye Hospital, Wenzhou Medical University, Wenzhou, China. 87Department of Ophthalmology and Visual Sciences, John Moran EyeCenter, University of Utah, Salt Lake City, UT, USA. 88State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University,Guangzhou, China. 89Translational Research Institute, University of Queensland Diamantina Institute, Brisbane, Queensland, Australia. 90MRC IntegrativeEpidemiology Unit, University of Bristol, Bristol, UK. 91Department of Pediatric Ophthalmology, Duke Eye Center For Human Genetics, Durham, NC, USA. 92Princess Alexandra Eye Pavilion, Edinburgh, UK. 93University Hospital ‘San Giovanni di Dio’, Cagliari, Italy. 94Department of Statistical Science, Schoolof Mathematics, Sun Yat-Sen University, Guangzhou, China. 95School of Social and Community Medicine, University of Bristol, Bristol, UK. 96Departmentof Clinical Physiology, Tampere University Hospital and School of Medicine, University of Tampere, Tampere, Finland. 97Institute for Molecular MedicineFinland FIMM, HiLIFE Unit, University of Helsinki, Helsinki, Finland. 98Division of Human Genetics, Genome Institute of Singapore, Singapore, Singapore. 99MRC Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King’s College London, London, UK. 100Université de Bordeaux, Bordeaux, France. 101Institut National de la Santé Et de la Recherche Médicale (INSERM), Institut de Santé Publiqued’Épidémiologie et de Développement (ISPED), Centre INSERM U897–Epidemiologie-Biostatistique, Bordeaux, France. 102Department of Ophthalmology, Yokohama City University School of Medicine, Yokohama, Japan. 103Suraj Eye Institute, Nagpur, Maharashtra, India. 104Institute of Neurogenomics, Helmholtz Zentrum München, German Research Centre for Environmental Health, Neuherberg, Germany. 105Institute of Genetic and Biomedic Research, National Research Council, Cagliari, Italy. 106Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands. 107MRC Epidemiology Unit, Instituteof Metabolic Sciences, University of Cambridge, Cambridge, UK. 108Centre for Myopia Research, School of Optometry, Hong Kong Polytechnic University, Hong Kong, Hong Kong. 109Department of Health Technology and Informatics, Hong Kong Polytechnic University, Hong Kong, Hong Kong. 110ManchesterRoyal Eye Hospital, Manchester University NHS Foundation Trust, Manchester Academic Health Science Centre, Manchester, UK. 111School of ComputerScience and Mathematics, Kingston University, Surrey, UK. 112Section of Epidemiology and Biostatistics, Leeds Institute of Cancer and Pathology, Universityof Leeds, Leeds, UK. 113Primary Care & Public Health Sciences, King’s College London, London, UK. 114Faculty of Medicine University of Southampton, Southampton General Hospital, Southampton, UK. 115School of Medicine, Dentistry and Biomedical Sciences, Queen’s University Belfast, Belfast, NorthernIreland, UK. 116Optometry and Visual Science, School of Health Science, City, University of London, London, UK. 117Division of Health Sciences & Centrefor Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK. 118School of Clinical Sciences, Faculty of Medicine and Dentistry, University of Bristol,Bristol, UK. 119Department of Psychiatry, Oxford University, Warneford Hospital, Oxford, UK. 120Department of Eye and Vision Science, University ofLiverpool, Liverpool, UK. 121Centre for Experimental Medicine, Queen’s University Belfast, Belfast, Northern Ireland, UK. 122Department of Ophthalmology, University of Southampton NHS Foundation Trust, Southampton, UK. 123Edinburgh Imaging, University of Edinburgh, Edinburgh, UK. 124Leeds Instituteof Rheumatic and Musculoskeletal Medicine, University of Leeds, Leeds, UK. 125Department of Ophthalmology, Cambridge University Hospitals NHSFoundation Trust, Cambridge, UK. 126Centre for Public Health, Queen’s University Belfast, Belfast, Northern Ireland, UK. 127Department of Ophthalmology,Leeds Teaching Hospitals NHS Trust, Leeds, UK. 128Department of Ophthalmology, King’s College Hospital NHS Foundation Trust, London, UK. 129StGeorge’s, University of London, London, UK. 130UCL Institute of Neurology, London, UK. 131Clinical and Experimental Sciences, Faculty of Medicine,University of Southampton, Southampton, UK. 132Institute of Genetic Medicine, Newcastle University, Newcastle Upon Tyne, UK. 133Gloucestershire RetinalResearch Group, Gloucestershire Hospitals NHS Foundation Trust, Cheltenham General Hospital, Cheltenham, UK. 134Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK. 135School of Science and Engineering, University of Dundee, Dundee, UK. 136Nottingham University Hospitals NHSTrust, Nottingham, UK. 137Norwich Medical School, University of East Anglia, Norwich, Norfolk, UK.
Rights and permissions
About this article
Cite this article
Tedja, M.S., Wojciechowski, R., Hysi, P.G. et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet 50, 834–848 (2018). https://doi.org/10.1038/s41588-018-0127-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0127-7
This article is cited by
-
The hazards of genotype imputation when mapping disease susceptibility variants
Genome Biology (2024)
-
Nutritional intake, environmental factors, and their impact on myopia prevalence in Korean children aged 5–12 years
Journal of Health, Population and Nutrition (2024)
-
Dissecting the complex sex-based associations of myopia with height and weight
Eye (2024)
-
POU6F2, a risk factor for glaucoma, myopia and dyslexia, labels specific populations of retinal ganglion cells
Scientific Reports (2024)
-
Diurnal retinal and choroidal gene expression patterns support a role for circadian biology in myopia pathogenesis
Scientific Reports (2024)