Abstract
Huntington disease (HD)-like 2 (HDL2) is a rare genetic disease caused by an expanded trinucleotide repeat in the JPH3 gene (encoding junctophilin 3) that shows remarkable clinical similarity to HD. To date, HDL2 has been reported only in patients with definite or probable African ancestry. A single haplotype background is shared by patients with HDL2 from different populations, supporting a common African origin for the expansion mutation. Nevertheless, outside South Africa, reports of patients with HDL2 in Africa are scarce, probably owing to limited clinical services across the continent. Systematic comparisons of HDL2 and HD have revealed closely overlapping motor, cognitive and psychiatric features and similar patterns of cerebral and striatal atrophy. The pathogenesis of HDL2 remains unclear but it is proposed to occur through several mechanisms, including loss of protein function and RNA and/or protein toxicity. This Review summarizes our current knowledge of this African-specific HD phenocopy and highlights key areas of overlap between HDL2 and HD. Given the aforementioned similarities in clinical phenotype and pathology, an improved understanding of HDL2 could provide novel insights into HD and other neurodegenerative and/or trinucleotide repeat expansion disorders.
Key points
-
Huntington disease (HD)-like 2 (HDL2) is a rare autosomal dominant genetic disease caused by a CTG/CAG trinucleotide repeat expansion in a variably spliced exon of the JPH3 gene (encoding junctophilin 3) located on chromosome 16q24.2.
-
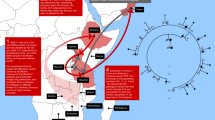
All documented patients with HDL2 have African ancestry and a shared haplotype across the JPH3 locus, suggesting a common ancient African origin, possibly in West Africa.
-
Fewer than 100 cases of HDL2 have been reported worldwide, emphasizing the need for concerted efforts to systematically ascertain patients for longitudinal studies.
-
HDL2 is the HD phenocopy that has the strongest clinical resemblance to HD across the disease course, with overlapping movement, psychiatric, cognitive and radiological features, progressing to non-verbal and akinetic dementia and premature death.
-
Loss of function of the junctophilin 3 protein, RNA toxicity of the sense strand and expanded CAG, and polyglutamine toxicity from the antisense strand have all been implicated in HDL2 pathogenesis.
-
Studies comparing HDL2 to HD provide unique opportunities to improve our understanding of the role of junctophilin 3 in the brain and of repeat expansion pathogenesis, allowing the development of novel biomarkers and therapeutic options.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Walker, F. O. Huntington’s disease. Lancet 369, 218–228 (2007).
Franklin, G. L., Teive, H. A. G., Meira, A. T., Nepomuceno, A. M. T. & Tabrizi, S. J. “On chorea”: 150 years of the beginning of hope. Mov. Disord. 37, 2194–2196 (2022).
MacDonald, M. E. et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983 (1993).
Margolis, R. L. et al. A disorder similar to Huntington’s disease is associated with a novel CAG repeat expansion. Ann. Neurol. 50, 373–380 (2001).
Margolis, R. L. & Rudnicki, D. D. Pathogenic insights from Huntington’s disease-like 2 and other Huntington’s disease genocopies. Curr. Opin. Neurol. 29, 743–748 (2016).
Krause, A. et al. Junctophilin 3 (JPH3) expansion mutations causing Huntington disease like 2 (HDL2) are common in South African patients with African ancestry and a Huntington disease phenotype. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 573–585 (2015).
Anderson, D. G. et al. Comparison of the Huntington’s disease like 2 and Huntington’s disease clinical phenotypes. Mov. Disord. Clin. Pract. 6, 302–311 (2019).
Tabrizi, S. J. et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 8, 791–801 (2009).
Paulsen, J. S. et al. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry 79, 874–880 (2008).
Ruderfer, D. M. & Dudley, J. T. Deep phenotyping predicts Huntington’s genotype. Nat. Biotechnol. 34, 823–824 (2016).
Weir, D. W., Sturrock, A. & Leavitt, B. R. Development of biomarkers for Huntington’s disease. Lancet Neurol. 10, 573–590 (2011).
Tabrizi, S. J. et al. Potential disease-modifying therapies for Huntington’s disease: lessons learned and future opportunities. Lancet Neurol. 21, 645–658 (2022).
Medina, A., Mahjoub, Y., Shaver, L. & Pringsheim, T. Prevalence and incidence of Huntington’s disease: an updated systematic review and meta-analysis. Mov. Disord. 37, 2327–2335 (2022).
Schneider, S. A. & Bird, T. Huntington’s disease, Huntington’s disease look-alikes, and benign hereditary chorea: what’s new? Mov. Disord. Clin. Pract. 3, 342–354 (2016).
Andrew, S. E. et al. Huntington disease without CAG expansion: phenocopies or errors in assignment? Am. J. Hum. Genet. 54, 852–863 (1994).
Wild, E. J. et al. Huntington’s disease phenocopies are clinically and genetically heterogeneous. Mov. Disord. 23, 716–720 (2008).
Schneider, S. A., Walker, R. H. & Bhatia, K. P. The Huntington’s disease-like syndromes: what to consider in patients with a negative Huntington’s disease gene test. Nat. Clin. Pract. Neurol. 3, 517–525 (2007).
Nguyen, Q. T. R. et al. Combining literature review with a ground truth approach for diagnosing Huntington’s disease phenocopy. Front. Neurol. 13, 817753 (2022).
Hensman Moss, D. J. et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology 82, 292–299 (2014).
Moore, R. C. et al. Huntington disease phenocopy is a familial prion disease. Am. J. Hum. Genet. 69, 1385–1388 (2001).
Kambouris, M., Bohlega, S., Al-Tahan, A. & Meyer, B. F. Localization of the gene for a novel autosomal recessive neurodegenerative Huntington-like disorder to 4p15.3. Am. J. Hum. Genet. 66, 445–452 (2000).
Rawlins, M. D. et al. The prevalence of Huntington’s disease. Neuroepidemiology 46, 144–153 (2016).
Baine, F. K. et al. Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur. J. Hum. Genet. 21, 1120–1127 (2013).
Cubo, E. et al. The burden of movement disorders in Cameroon: a rural and urban-based inpatient/outpatient study. Mov. Disord. Clin. Pract. 4, 568–573 (2017).
Baine, F. K., Krause, A. & Greenberg, L. J. The frequency of Huntington disease and Huntington disease-like 2 in the South African population. Neuroepidemiology 46, 198–202 (2016).
Kay, C. et al. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 177, 346–357 (2018).
Holmes, S. E. et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat. Genet. 29, 377–378 (2001).
Margolis, R. L. et al. Huntington’s disease-like 2 (HDL2) in North America and Japan. Ann. Neurol. 56, 670–674 (2004).
Robbins, N. M. et al. Black patients matter in neurology: race, racism, and race-based neurodisparities. Neurology 99, 106–114 (2022).
Walker, R. H. et al. Huntington’s disease-like disorders in Latin America and the Caribbean. Parkinsonism Relat. Disord. 53, 10–20 (2018).
Rodrigues, G. G. R. et al. Huntington’s disease-like 2 in Brazil–report of 4 patients. Mov. Disord. 23, 2244–2247 (2008).
Paradisi, I., Ikonomu, V. & Arias, S. Huntington disease-like 2 Venezuela: frequency ethnic origin. J. Hum. Genet. 58, 3–6 (2013).
Santos, C. et al. Huntington disease-like 2: the first patient with apparent European ancestry. Clin. Genet. 73, 480–485 (2008).
Parra, F. C. et al. Color and genomic ancestry in Brazilians. Proc. Natl Acad. Sci. USA 100, 177–182 (2003).
de Souza, A. M., Resende, S. S., de Sousa, T. N. & de Brito, C. F. A. A systematic scoping review of the genetic ancestry of the Brazilian population. Genet. Mol. Biol. 42, 495–508 (2019).
Rodrigues, G. G. R., Teive, H. A. G. & Tumas, V. Huntington’s disease-like 2 and apparent ancestry. Clin. Genet. 75, 207 (2009).
Anderson, D. G. et al. A systematic review of the Huntington disease-like 2 phenotype. J. Huntingt. Dis. 6, 37–46 (2017).
Stevanin, G. et al. Huntington’s disease-like phenotype due to trinucleotide repeat expansions in the TBP and JPH3 genes. Brain 126, 1599–1603 (2003).
Mariani, L. et al. Expanding the spectrum of genes involved in Huntington disease using a combined clinical and genetic approach. JAMA Neurol. 73, 1105–1114 (2016).
Ruscitti, F. et al. A case of Huntington disease-like 2 in a patient of African ancestry: the everlasting support of clinical examination in the molecular era. Clin. Case Rep. 10, e6308 (2022).
Baine, F. K., Peerbhai, N. & Krause, A. A study of Huntington disease-like syndromes in black South African patients reveals a single SCA2 mutation and a unique distribution of normal alleles across five repeat loci. J. Neurol. Sci. 390, 200–204 (2018).
Ocampo, C., Daimari, R. & Oyekunle, A. A. Huntington’s disease-like 2 with an expansion mutation of the Junctophilin-3 gene; first reported case from Botswana. J. Clin. Neurosci. 47, 126–127 (2018).
Micheletti, S. J. et al. Genetic consequences of the transatlantic slave trade in the Americas. Am. J. Hum. Genet. 107, 265–277 (2020).
Muthinja, M. J. et al. An exploration of the genetics of the mutant Huntingtin (mHtt) gene in a cohort of patients with chorea from different tribes in 6 sub-Saharan African countries. Preprint at medRxiv https://doi.org/10.1101/2022.07.13.22272435 (2022).
Chukwuneke, F. N., Ezeonu, C. T., Onyire, B. N. & Ezeonu, P. O. Culture and biomedical care in Africa: the influence of culture on biomedical care in a traditional African society, Nigeria, West Africa. Niger. J. Med. 21, 331–333 (2012).
Glover, S. M. Mark Nichter: global health: why cultural perceptions, social representations, and biopolitics matter. Hum. Ecol. 37, 669–670 (2009).
Seixas, A. I. et al. Loss of junctophilin-3 contributes to Huntington disease-like 2 pathogenesis. Ann. Neurol. 71, 245–257 (2012).
Hayden, M. R., MacGregor, J. M., Saffer, D. S. & Beighton, P. H. The high frequency of juvenile Huntington’s chorea in South Africa. J. Med. Genet. 19, 94–97 (1982).
Quarrell, O., O’Donovan, K. L., Bandmann, O. & Strong, M. the prevalence of juvenile Huntington’s disease: a review of the literature and meta-analysis. PLoS Curr. 4, e4f8606b742ef3 (2012).
Bardien, S. et al. A South African mixed ancestry family with Huntington disease-like 2: clinical and genetic features. Mov. Disord. 22, 2083–2089 (2007).
Warby, S. C. et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 84, 351–366 (2009).
Pearson, C. E., Nichol Edamura, K. & Cleary, J. D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 (2005).
McMurray, C. T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11, 786–799 (2010).
Goldman, A., Ramsay, M. & Jenkins, T. Absence of myotonic dystrophy in southern African Negroids is associated with a significantly lower number of CTG trinucleotide repeats. J. Med. Genet. 31, 37–40 (1994).
Gennarelli, M. et al. CTG repeats distribution and Alu insertion polymorphism at myotonic dystrophy (DM) gene in Amhara and Oromo populations of Ethiopia. Hum. Genet. 105, 165–167 (1999).
Chiurazzi, P. et al. Extended gene diversity at the FMR1 locus and neighbouring CA repeats in a sub-Saharan population. Am. J. Med. Genet. 64, 216–219 (1996).
Ritchie, R. J., Chakrabarti, L., Knight, S. J., Harding, R. M. & Davies, K. E. Population genetics of the FRAXE and FRAXF GCC repeats, and a novel CGG repeat, in Xq28. Am. J. Med. Genet. 73, 463–469 (1997).
Peprah, E. K., Allen, E. G., Williams, S. M., Woodard, L. M. & Sherman, S. L. Genetic diversity of the fragile X syndrome gene (FMR1) in a large sub-Saharan West African population. Ann. Hum. Genet. 74, 316–325 (2010).
Levesley, J. Investigating allele sequence diversity at the Huntington disease loci HTT and JPH3 in African ancestry individuals. The University of the Witwatersrand: Electronic Theses and Dissertations https://wiredspace.wits.ac.za/items/25004bb1-ea69-4035-bec5-1134d077c768. (2021).
Ibañez, K. et al. Population frequency of repeat expansions indicates increased disease prevalence estimates across different populations. Preprint at: medRxiv https://doi.org/10.1101/2023.07.03.23292162 (2023).
Squitieri, F. et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum. Mol. Genet. 3, 2103–2114 (1994).
Ciosi, M. et al. A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. EBioMedicine 48, 568–580 (2019).
Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900.e14 (2019).
Wright, G. E. B. et al. Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am. J. Hum. Genet. 104, 1116–1126 (2019).
Dawson, J. et al. A probable cis-acting genetic modifier of Huntington disease frequent in individuals with African ancestry. HGG Adv. 3, 100130 (2022).
Maiuri, T. et al. DNA damage repair in Huntington’s disease and other neurodegenerative diseases. Neurotherapeutics 16, 948–956 (2019).
Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 162, 516–526 (2015).
Lee, J. et al. Genetic modifiers of Huntington disease differentially influence motor and cognitive domains. Am. J. Hum. Genet. 109, 885–899 (2022).
Greenstein, P. E., Vonsattel, J. G., Margolis, R. L. & Joseph, J. T. Huntington’s disease like-2 neuropathology. Mov. Disord. 22, 1416–1423 (2007).
Walker, R. H. et al. Autosomal dominant chorea-acanthocytosis with polyglutamine-containing neuronal inclusions. Neurology 58, 1031–1037 (2002).
Walker, R. H. et al. Huntington’s disease-like 2 can present as chorea-acanthocytosis. Neurology 61, 1002–1004 (2003).
Krause, A., Temlett, J. & Van der Meyden, K. CAG/CTG repeat expansions at the HDL2 locus are a common cause of Huntington disease in Black South Africans (Abstr.). Am. J. Hum. Genet. 71, 528 (2002).
Rudnicki, D. D., Pletnikova, O., Vonsattel, J. G., Ross, C. A. & Margolis, R. L. A comparison of Huntington disease and Huntington disease-like 2 neuropathology. J. Neuropathol. Exp. Neurol. 67, 366–374 (2008).
Anderson, D. G. et al. Emerging differences between Huntington’s disease-like 2 and Huntington’s disease: a comparison using MRI brain volumetry. Neuroimage Clin. 21, 101666 (2019).
Stevanin, G. et al. CAG/CTG repeat expansions at the Huntington’s disease-like 2 locus are rare in Huntington’s disease patients. Neurology 58, 965–967 (2002).
Schneider, S. A., Marshall, K. E., Xiao, J. & LeDoux, M. S. JPH3 repeat expansions cause a progressive akinetic-rigid syndrome with severe dementia and putaminal rim in a five-generation African-American family. Neurogenetics 13, 133–140 (2012).
Walker, R. H., Jankovic, J., O’Hearn, E. & Margolis, R. L. Phenotypic features of Huntington’s disease-like 2. Mov. Disord. 18, 1527–1530 (2003).
Mulroy, E. et al. Huntington disease like 2 (HDL-2) with parkinsonism and abnormal DAT-SPECT — a novel observation. Parkinsonism Relat. Disord. 71, 46–48 (2020).
[No authors listed] Unified Huntington’s disease rating scale: reliability and consistency. Huntington Study Group.Mov. Disord. 11, 136–142 (1996).
Lasker, A. G. & Zee, D. S. Ocular motor abnormalities in Huntington’s disease. Vis. Res. 37, 3639–3645 (1997).
Rosenblatt, A. et al. Age, CAG repeat length, and clinical progression in Huntington’s disease. Mov. Disord. 27, 272–276 (2012).
Rudnicki, D. D. et al. Huntington’s disease-like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 61, 272–282 (2007).
Ferreira-Correia, A., Krause, A. & Anderson, D. G. The neuropsychiatry of Huntington disease-like 2: a comparison with Huntington’s disease. J. Huntingt. Dis. 9, 325–334 (2020).
Margolis, R. L. & Holmes, S. E. Huntington’s disease-like 2: a clinical, pathological, and molecular comparison to Huntington’s disease. Clin. Neurosci. Res. 3, 187–196 (2003).
Duff, K. et al. Psychiatric symptoms in Huntington’s disease before diagnosis: the predict-HD study. Biol. Psychiatry 62, 1341–1346 (2007).
Fischer, C. A., Licht, E. A. & Mendez, M. F. The neuropsychiatric manifestations of Huntington’s disease-like 2. J. Neuropsychiatry Clin. Neurosci. 24, 489–492 (2012).
Castilhos, R. M. et al. Huntington disease and Huntington disease-like in a case series from Brazil. Clin. Genet. 86, 373–377 (2014).
Paradisi, I., Ikonomu, V. & Arias, S. Huntington disease-like 2 (HDL2) in Venezuela: frequency and ethnic origin. J. Hum. Genet. 58, 3–6 (2013).
Ferreira-Correia, A., Anderson, D. G., Cockcroft, K. & Krause, A. The neuropsychological deficits and dissociations in Huntington disease-like 2: a series of case-control studies. Neuropsychologia 136, 107238 (2020).
Ferreira-Correia, A., Anderson, D. G., Cockcroft, K. & Krause, A. A comparison between the neurocognitive profile of Huntington disease-like 2 and Huntington disease: exploring the presence of double dissociations. Appl. Neuropsychol. Adult 29, 223–233 (2022).
Ferreira-Correia, A. & Cockcroft, K. Controlling for inequality in neuropsychological assessment: using Crawford and Howell’s (1998) single-case methodology with norms from demographically homogeneous groups of South Africans. S. Afr. J. Psychol.53, https://doi.org/10.1177/0081246322115100 (2023).
Roussakis, A. & Piccini, P. PET imaging in Huntington’s disease. J. Huntingt. Dis. 4, 287–296 (2015).
Gamez, J. et al. Does reduced [123I]-FP-CIT binding in Huntington’s disease suggest pre-synaptic dopaminergic involvement? Clin. Neurol. Neurosurg. 112, 870–875 (2010).
Trottier, Y. et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature 378, 403–406 (1995).
Danek, A. & Walker, R. H. Neuroacanthocytosis. Curr. Opin. Neurol. 18, 386–392 (2005).
Anderson, D. G. et al. Absence of acanthocytosis in Huntington’s disease-like 2: a prospective comparison with Huntington’s disease. Tremor Other Hyperkinet Mov. 7, 512 (2017).
Walker, R. H. & Danek, A. “Neuroacanthocytosis” — overdue for a taxonomic update. Tremor Other Hyperkinet Mov. 11, 1 (2021).
Tabrizi, S. J., Flower, M. D., Ross, C. A. & Wild, E. J. Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 16, 529–546 (2020).
Iyer, R. R. & Pluciennik, A. DNA mismatch repair and its role in Huntington’s disease. J. Huntingt. Dis. 10, 75–94 (2021).
Marti, E. RNA toxicity induced by expanded CAG repeats in Huntington’s disease. Brain Pathol. 26, 779–786 (2016).
Banez-Coronel, M. et al. RAN translation in Huntington disease. Neuron 88, 667–677 (2015).
Sathasivam, K. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA 110, 2366–2370 (2013).
Chung, D. W., Rudnicki, D. D., Yu, L. & Margolis, R. L. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477 (2011).
Saudou, F. & Humbert, S. The biology of huntingtin. Neuron 89, 910–926 (2016).
Lehnart, S. E. & Wehrens, X. H. T. The role of junctophilin proteins in cellular function. Physiol. Rev. 102, 1211–1261 (2022).
Piggott, C. A. et al. Caenorhabditis elegans junctophilin has tissue-specific functions and regulates neurotransmission with extended-synaptotagmin. Genetics 218, iyab063 (2021).
Takeshima, H., Komazaki, S., Nishi, M., Iino, M. & Kangawa, K. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11–22 (2000).
Landstrom, A. P., Beavers, D. L. & Wehrens, X. H. T. The junctophilin family of proteins: from bench to bedside. Trends Mol. Med. 20, 353–362 (2014).
Kakizawa, S., Moriguchi, S., Ikeda, A., Iino, M. & Takeshima, H. Functional crosstalk between cell-surface and intracellular channels mediated by junctophilins essential for neuronal functions. Cerebellum 7, 385–391 (2008).
Takeshima, H., Hoshijima, M. & Song, L. Ca2+ microdomains organized by junctophilins. Cell Calcium 58, 349–356 (2015).
Sahu, G. et al. Junctophilin proteins tether a Cav1-RyR2-KCa3.1 tripartite complex to regulate neuronal excitability. Cell. Rep. 28, 2427–2442.e6 (2019).
Guo, A. et al. E-C coupling structural protein junctophilin-2 encodes a stress-adaptive transcription regulator. Science 362, eaan3303 (2018).
Lahiri, S. K. et al. Nuclear localization of a novel calpain-2 mediated junctophilin-2 C-terminal cleavage peptide promotes cardiomyocyte remodeling. Basic Res. Cardiol. 115, 49 (2020).
Nishi, M. et al. Motor discoordination in mutant mice lacking junctophilin type 3. Biochem. Biophys. Res. Commun. 292, 318–324 (2002).
Calpena, E. et al. The Drosophila junctophilin gene is functionally equivalent to its four mammalian counterparts and is a modifier of a Huntingtin poly-Q expansion and the Notch pathway. Dis. Model. Mech. 11, dmm029082 (2018).
Firth, H. V. et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009).
Bourinaris, T. et al. Allelic and phenotypic heterogeneity in Junctophillin-3 related neurodevelopmental and movement disorders. Eur. J. Hum. Genet. 29, 1027–1031 (2021).
Steel, D. et al. Both heterozygous and homozygous loss-of-function JPH3 variants are associated with a paroxysmal movement disorder. Mov. Disord. 38, 155–157 (2023).
Auerbach, W. et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet. 10, 2515–2523 (2001).
Murthy, V. et al. Hypomorphic mutation of the mouse Huntington’s disease gene orthologue. PLoS Genet. 15, e1007765 (2019).
Timchenko, L. Development of therapeutic approaches for myotonic dystrophies type 1 and type 2. Int. J. Mol. Sci. 23, 10491 (2022).
Wilburn, B. et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington’s disease-like 2 mice. Neuron 70, 427–440 (2011).
Nucifora, F. C. J. et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291, 2423–2428 (2001).
Rodrigues, G. R. et al. Clinical and genetic analysis of 29 Brazilian patients with Huntington’s disease-like phenotype. Arq. Neuropsiquiatr. 69, 419–423 (2011).
Bonomo, R. et al. Deep brain stimulation in Huntington’s disease: a literature review. Neurol. Sci. 42, 4447–4457 (2021).
Yu, H., Takahashi, K., Bloom, L., Quaynor, S. D. & Xie, T. Effect of deep brain stimulation on swallowing function: a systematic review. Front. Neurol. 11, 547 (2020).
Testa, C. M. & Jankovic, J. Huntington disease: a quarter century of progress since the gene discovery. J. Neurol. Sci. 396, 52–68 (2019).
Jiang, J., Tang, M., Huang, Z. & Chen, L. Junctophilins emerge as novel therapeutic targets. J. Cell. Physiol. 234, 16933–16943 (2019).
Sathe, S. et al. Enroll-HD: an integrated clinical research platform and worldwide observational study for Huntington’s disease. Front. Neurol. 12, 667420 (2021).
Magazi, D. S. et al. Huntington’s disease: genetic heterogeneity in black African patients. S. Afr. Med. J. 98, 200–203 (2008).
Vasconcellos, L. F. R. et al. Huntington’s Disease like 2 presenting with isolated Parkinsonism. J. Neurol. Sci. 373, 105–106 (2017).
Teive, H. A. G. et al. Huntington’s disease-like 2: the first case report in Latin America in a patient without African ethnic origin. Mov. Disord. 22, S27 (2007).
Seeley, A. H. et al. Macrocerebellum, epilepsy, intellectual disability, and gut malrotation in a child with a 16q24.1-q24.2 contiguous gene deletion. Am. J. Med. Genet. A 164A, 2062–2068 (2014).
Perni, S. & Beam, K. Neuronal junctophilins recruit specific CaV and RyR isoforms to ER-PM junctions and functionally alter CaV2.1 and CaV2.2. eLife 10, e64249 (2021).
Author information
Authors and Affiliations
Contributions
A.K., D.G.A., A.F.-C. and R.L.M. researched data for the article and made substantial contributions to discussions of the content, and each wrote a section. J.D. and F.B.-S. contributed to the Genetics section. J.D. generated some of the unpublished data mentioned in the article. P.P.L. contributed to the Pathogenesis section and references. All authors edited and reviewed the document before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Neurology thanks R. Walker, S. Schneider and E. Wild for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Acanthocytes
-
Red blood cells with spiculated protrusions from the cell membrane of varying size and distribution.
- Anticipation
-
A phenomenon whereby disease onset occurs earlier and more severely with each successive generation.
- Synonymous sequence variation
-
DNA sequence variation that does not change the amino acid sequence of the encoded protein.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Krause, A., Anderson, D.G., Ferreira-Correia, A. et al. Huntington disease-like 2: insight into neurodegeneration from an African disease. Nat Rev Neurol 20, 36–49 (2024). https://doi.org/10.1038/s41582-023-00906-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41582-023-00906-y