Abstract
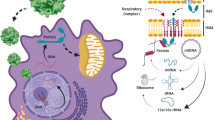
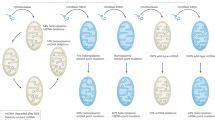
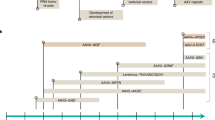
The variable clinical and biochemical manifestations of primary mitochondrial diseases (PMDs), and the complexity of mitochondrial genetics, have proven to be a substantial barrier to the development of effective disease-modifying therapies. Encouraging data from gene therapy trials in patients with Leber hereditary optic neuropathy and advances in DNA editing techniques have raised expectations that successful clinical transition of genetic therapies for PMDs is feasible. However, obstacles to the clinical application of genetic therapies in PMDs remain; the development of innovative, safe and effective genome editing technologies and vectors will be crucial to their future success and clinical approval. In this Perspective, we review progress towards the genetic treatment of nuclear and mitochondrial DNA-related PMDs. We discuss advances in mitochondrial DNA editing technologies alongside the unique challenges to targeting mitochondrial genomes. Last, we consider ongoing trials and regulatory requirements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Al-Zaidy, S. et al. Health outcomes in spinal muscular atrophy type 1 following AVXS-101 gene replacement therapy. Pediatr. Pulmonol. 54, 179–185 (2019).
Russell, S. et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017).
Thompson, A. A. et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N. Engl. J. Med. 378, 1479–1493 (2018).
Kuzmin, D. A. et al. The clinical landscape for AAV gene therapies. Nat. Rev. Drug. Discov. 20, 173–174 (2021).
Iftikhar, M., Frey, J., Shohan, M. J., Malek, S. & Mousa, S. A. Current and emerging therapies for Duchenne muscular dystrophy and spinal muscular atrophy. Pharmacol. Ther. 220, 107719 (2021).
Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
Gorman, G. S. et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 (2015).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016).
Pitceathly, R. D. S., Keshavan, N., Rahman, J. & Rahman, S. Moving towards clinical trials for mitochondrial diseases. J. Inherit. Metab. Dis. 44, 22–41 (2021).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res 49, D1541–D1547 (2021).
Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981).
Gustafsson, C. M., Falkenberg, M. & Larsson, N. G. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 (2016).
Ng, Y. S. et al. Mitochondrial disease in adults: recent advances and future promise. Lancet Neurol. 20, 573–584 (2021).
Schon, E. A., Dimauro, S. & Hirano, M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878–890 (2012).
Larsson, N. G. & Clayton, D. A. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29, 151–178 (1995).
Rossignol, R. et al. Mitochondrial threshold effects. Biochem. J. 370, 751–762 (2003).
Bugiardini, E. et al. Expanding the molecular and phenotypic spectrum of truncating MT-ATP6 mutations. Neurol. Genet. 6, e381 (2020).
Pitceathly, R. D. S. et al. Genetic dysfunction of MT-ATP6 causes axonal Charcot-Marie-Tooth disease. Neurology 79, 1145–1154 (2012).
Banskota, S. et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 185, 250–265.e16 (2022).
Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384, 252–260 (2021).
Ribeil, J.-A. et al. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med. 376, 848–855 (2017).
Mendell, J. R. et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713–1722 (2017).
Lundstrom, K. Viral vectors in gene therapy. Diseases 6, 42 (2018).
Crooke, S. T., Baker, B. F., Crooke, R. M. & Liang, X. H. Antisense technology: an overview and prospectus. Nat. Rev. Drug. Discov. 20, 427–453 (2021).
Silva-Pinheiro, P. & Minczuk, M. The potential of mitochondrial genome engineering. Nat. Rev. Genet. 23, 199–214 (2022).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Stewart, J. B. Current progress with mammalian models of mitochondrial DNA disease. J. Inherit. Metab. Dis. 44, 325–342 (2021).
King, M. P. & Attardi, G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503 (1989).
Brown, M. D., Trounce, I. A., Jun, A. S., Allen, J. C. & Wallace, D. C. Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber’s hereditary optic neuropathy mitochondrial DNA mutation. J. Biol. Chem. 275, 39831–39836 (2000).
Pallotti, F. et al. Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. Biochem. J. 384, 287–293 (2004).
Trounce, I., Neill, S. & Wallace, D. C. Cytoplasmic transfer of the mtDNA nt 8993 T->G (ATP6) point mutation associated with Leigh syndrome into mtDNA-less cells demonstrates cosegregation with a decrease in state III respiration and ADP/O ratio. Proc. Natl Acad. Sci. USA 91, 8334–8338 (1994).
Cavaliere, A., Marchet, S., Di Meo, I. & Tiranti, V. An in vitro approach to study mitochondrial dysfunction: a cybrid model. J. Vis. Exp. 181, e63452 (2022).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Wang, D., L Tai, P. W. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug. Discov. 18, 358–378 (2019).
Torres-Torronteras, J. et al. Long-term sustained effect of liver-targeted adeno-associated virus gene therapy for mitochondrial neurogastrointestinal encephalomyopathy. Hum. Gene Ther. 29, 708–718 (2018).
Vila-Julià, F. et al. Efficacy of adeno-associated virus gene therapy in a MNGIE murine model enhanced by chronic exposure to nucleosides. EBioMedicine 62, 103133 (2020).
Torres-Torronteras, J. et al. Gene therapy using a liver-targeted AAV vector restores nucleoside and nucleotide homeostasis in a murine model of MNGIE. Mol. Ther. 22, 901–907 (2014).
Bottani, E. et al. AAV-mediated liver-specific MPV17 expression restores mtDNA levels and prevents diet-induced liver failure. Mol. Ther. 22, 10–17 (2014).
Bacman, S. R. et al. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. 24, 1696 (2018).
Gammage, P. A. et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med. 24, 1691–1695 (2018).
Di Meo, I. et al. Effective AAV-mediated gene therapy in a mouse model of ethylmalonic encephalopathy. EMBO Mol. Med. 4, 1008 (2012).
Wang, Z. et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 23, 321–328 (2005).
Silva-Pinheiro, P., Cerutti, R., Luna-Sanchez, M., Zeviani, M. & Viscomi, C. A single intravenous injection of AAV-PHP.B-hNDUFS4 ameliorates the phenotype of Ndufs4−/− mice. Mol. Ther. Methods Clin. Dev. 17, 1071–1078 (2020).
Yang, L. et al. Systemic administration of AAV-Slc25a46 mitigates mitochondrial neuropathy in Slc25a46−/− mice. Hum. Mol. Genet 29, 649–661 (2020).
Ling, Q., Rioux, M., Hu, Y., Lee, M. J. & Gray, S. J. Adeno-associated viral vector serotype 9-based gene replacement therapy for SURF1-related Leigh syndrome. Mol. Ther. Methods Clin. Dev. 23, 158–168 (2021).
Lopez-Gomez, C. et al. Synergistic deoxynucleoside and gene therapies for thymidine kinase 2 deficiency. Ann. Neurol. 90, 640–652 (2021).
Reynaud-Dulaurier, R. et al. Gene replacement therapy provides benefit in an adult mouse model of Leigh syndrome. Brain 143, 1686–1696 (2020).
Corrà, S., Cerutti, R., Balmaceda, V., Viscomi, C.& Zeviani, M. Double administration of self-complementary AAV9NDUFS4 prevents Leigh disease in Ndufs4−/− mice. Brain 139, 16–17 (2022).
Dong, J. Y., Fan, P. D. & Frizzell, R. A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 7, 2101–2112 (1996).
Rahman, S. & Copeland, W. C. POLG-related disorders and their neurological manifestations. Nat. Rev. Neurol. 15, 40–52 (2018).
Saneto, R. P. & Naviaux, R. K. Polymerase gamma disease through the ages. Dev. Disabil. Res. Rev. 16, 163–174 (2010).
Tornabene, P. et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 11, eaav4523 (2019).
Tornabene, P. & Trapani, I. Can adeno-associated viral vectors deliver effectively large genes? Hum. Gene Ther. 31, 47–56 (2020).
Zhi, S. et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 30, 283–294 (2022).
Bennett, C. F., Baker, B. F., Pham, N., Swayze, E. & Geary, R. S. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 57, 81–105 (2017).
Venkatesh, A. et al. Antisense oligonucleotide mediated increase of OPA1 expression using TANGO technology for the treatment of autosomal dominant optic atrophy [abstract]. Investig. Ophthalmol. Vis. Sci. 61, 2755 (2020).
Indrieri, A. et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med. 11, e8734 (2019).
Bacman, S. R., Williams, S. L., Pinto, M. & Moraes, C. T. The use of mitochondria-targeted endonucleases to manipulate mtDNA. Methods Enzymol. 547, 373–397 (2014).
Bacman, S. R., Williams, S. L., Pinto, M., Peralta, S. & Moraes, C. T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 19, 1111–1113 (2013).
Gammage, P. A., Rorbach, J., Vincent, A. I., Rebar, E. J. & Minczuk, M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med. 6, 458–466 (2014).
Zekonyte, U. et al. Mitochondrial targeted meganuclease as a platform to eliminate mutant mtDNA in vivo. Nat. Commun. 12, 3210 (2021).
Gammage, P. A., Moraes, C. T. & Minczuk, M. Mitochondrial genome engineering: the revolution may not be CRISPR-ized. Trends Genet 34, 101–110 (2018).
Tanaka, M. et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J. Biomed. Sci. 9, 534–541 (2002).
Alexeyev, M. F. et al. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 15, 516–523 (2008).
Bayona-Bafaluy, M. P., Blits, B., Battersby, B. J., Shoubridge, E. A. & Moraes, C. T. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc. Natl Acad. Sci. USA 102, 14392–14397 (2005).
Bacman, S. R., Williams, S. L., Duan, D. & Moraes, C. T. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 19, 1101–1106 (2012).
Gammage, P. A. et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res 44, 7804–7816 (2016).
Jackson, C. B., Turnbull, D. M., Minczuk, M. & Gammage, P. A. Therapeutic manipulation of mtDNA heteroplasmy: a shifting perspective. Trends Mol. Med. 26, 698–709 (2020).
Minczuk, M., Papworth, M. A., Miller, J. C., Murphy, M. P. & Klug, A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 36, 3926–3938 (2008).
Gaude, E. et al. NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol. Cell 69, 581–593.e7 (2018).
Yang, Y. et al. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell 9, 283–297 (2018).
Yahata, N., Boda, H. & Hata, R. Elimination of mutant mtDNA by an optimized mpTALEN restores differentiation capacities of heteroplasmic MELAS-iPSCs. Mol. Ther. Methods Clin. Dev. 20, 54–68 (2020).
Hashimoto, M. et al. MitoTALEN: a general approach to reduce mutant mtDNA loads and restore oxidative phosphorylation function in mitochondrial diseases. Mol. Ther. 23, 1592–1599 (2015).
Reddy, P. et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell 161, 459–469 (2015).
Pereira, C. V. et al. mitoTev-TALE: a monomeric DNA editing enzyme to reduce mutant mitochondrial DNA levels. EMBO Mol. Med. 10, e8084 (2018).
Arnould, S. et al. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng. Des. Sel. 24, 27–31 (2011).
Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020).
Cho, S.-I. et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell 185, 1764–1776 (2022).
Lei, Z. et al. Mitochondrial base editor induces substantial nuclear off-target mutations. Nature 606, 804–811 (2022).
Man, P. Y. W. et al. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am. J. Hum. Genet. 72, 333–339 (2003).
Yu-Wai-Man, P. et al. A neurodegenerative perspective on mitochondrial optic neuropathies. Acta Neuropathol. 132, 789 (2016).
Wallace, D. C. et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242, 1427–1430 (1988).
Yu, H. et al. Gene delivery to mitochondria by targeting modified adenoassociated virus suppresses Leber’s hereditary optic neuropathy in a mouse model. Proc. Natl Acad. Sci. USA 109, 1238–1247 (2012).
Mingozzi, F. & High, K. A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood 122, 23–36 (2013).
Peverelli, L. et al. Leber’s hereditary optic neuropathy: a report on novel mtDNA pathogenic variants. Front. Neurol. 12, 652 (2021).
Yu-Wai-Man, P. et al. Bilateral visual improvement with unilateral gene therapy injection for Leber hereditary optic neuropathy. Sci. Transl. Med. 12, eaaz7423 (2020).
Newman, N. J. et al. Efficacy and safety of intravitreal gene therapy for Leber hereditary optic neuropathy treated within 6 months of disease onset. Ophthalmology 128, 649–660 (2021).
Newman, N. J. et al. Intravitreal gene therapy vs. natural history in patients with Leber hereditary optic neuropathy carrying the m.11778G>A ND4 mutation: systematic review and indirect comparison. Front. Neurol. 12, 662838 (2021).
Biousse, V. et al. Long-term follow-up after unilateral intravitreal gene therapy for Leber hereditary optic neuropathy: the RESTORE study. J. Neuroophthalmol. 41, 309–315 (2021).
Bonnet, C. et al. The optimized allotopic expression of ND1 or ND4 genes restores respiratory chain complex I activity in fibroblasts harboring mutations in these genes. Biochim. Biophys. Acta 1783, 1707–1717 (2008).
Koilkonda, R. et al. LHON gene therapy vector prevents visual loss and optic neuropathy induced by G11778A mutant mitochondrial DNA: biodistribution and toxicology profile. Investig. Ophthalmol. Vis. Sci. 55, 7739–7753 (2014).
Calkins, D. J. et al. Biodistribution of intravitreal lenadogene nolparvovec gene therapy in nonhuman primates. Mol. Ther. Methods Clin. Dev. 23, 307 (2021).
Perales-Clemente, E., Fernández-Silva, P., Acín-Pérez, R., Pérez-Martos, A. & Enríquez, J. A. Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res 39, 225–234 (2011).
Lewis, C. J. et al. Codon optimization is an essential parameter for the efficient allotopic expression of mtDNA genes. Redox Biol. 30, 101429 (2020).
Oca-Cossio, J., Kenyon, L., Hao, H. & Moraes, C. T. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics 165, 707–720 (2003).
Goto, Y. I., Nonaka, I. & Horai, S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 (1990).
El-Hattab, A. W., Almannai, M. & Scaglia, F. in Gene Reviews (Univ. Washington, 2018).
Lawlor, M. W. & Dowling, J. J. X-linked myotubular myopathy. Neuromuscul. Disord. 31, 1004–1012 (2021).
Bessis, N., GarciaCozar, F. J. & Boissier, M. C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 11, S10–S17 (2004).
Chan, Y. K. et al. Engineering adeno-associated viral vectors to evade innate immune and inflammatory responses. Sci. Transl. Med. 13, eabd3438 (2021).
Tiku, V., Tan, M. W. & Dikic, I. Mitochondrial functions in infection and immunity. Trends Cell Biol. 30, 263–275 (2020).
Suomalainen, A. et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 10, 806–818 (2011).
Maresca, A. et al. Expanding and validating the biomarkers for mitochondrial diseases. J. Mol. Med. 98, 1467–1478 (2020).
Lehtonen, J. M. et al. FGF21 is a biomarker for mitochondrial translation and mtDNA maintenance disorders. Neurology 87, 2290–2299 (2016).
Yatsuga, S. et al. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann. Neurol. 78, 814–823 (2015).
Parikh, S. et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet. Med. 17, 689 (2015).
Bulcha, J. T., Wang, Y., Ma, H., Tai, P. W. L. & Gao, G. Viral vector platforms within the gene therapy landscape. Signal. Transduct. Target. Ther. 6, 53 (2021).
Pereira, C. V. et al. Myopathy reversion in mice after restauration of mitochondrial complex I. EMBO Mol. Med. 12, e10674 (2020).
Yamada, Y., Somiya, K., Miyauchi, A., Osaka, H. & Harashima, H. Validation of a mitochondrial RNA therapeutic strategy using fibroblasts from a Leigh syndrome patient with a mutation in the mitochondrial ND3 gene. Sci. Rep. 10, 7511 (2020).
Yamada, Y. et al. Power of mitochondrial drug delivery systems to produce innovative nanomedicines. Adv. Drug. Deliv. Rev. 154–155, 187–209 (2020).
Holt, I. J., Harding, A. E. & Morgan-Hughes, J. A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 (1988).
Srivastava, S. & Moraes, C. T. Manipulating mitochondrial DNA heteroplasmy by mitochondrially targeted restriction endonuclease. Hum. Mol. Genet. 10, 3093–3099 (2001).
Silva-Pinheiro, P. et al. In vivo mitochondrial base editing via adeno-associated viral delivery to mouse post-mitotic tissue. Nat. Commun. 13, 750 (2022).
Acknowledgements
C.V. is supported by the Telethon Foundation (GGP20013), AFM-Telethon (23706), Associazione Luigi Comini Onlus, Department of Biomedical Sciences-UNIPD (SID2022- VISC_BIRD2222_01). M.M. is supported by the Medical Research Council (MC_UU_00028/3). R.D.S.P. and M.F. are supported by a Medical Research Council (UK) Clinician Scientist Fellowship (MR/S002065/1). M.F., M.M., M.G.H, C.V., and R.D.S.P. are supported by Medical Research Council (UK) award MC_PC_21046 to establish a National Mouse Genetics Network Cluster in Mitochondria (MitoCluster). M.G.H. and R.D.S.P. are supported by Medical Research Council (UK) strategic award MR/S005021/1 to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
M.M. is a co-founder, shareholder, and member of the Scientific Advisory Board of Pretzel Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Neurology thanks Robert Lightowlers, Ramon Martí, Yuma Yamada and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Cybrid
-
A cytoplasmic hybrid created by fusing cells harbouring a wild-type or altered mtDNA of interest with cells depleted of endogenous mtDNA.
- Heteroplasmy shift
-
A shift in the relative abundance of mutant mtDNA to wild-type mtDNA.
- Indels
-
The insertion and/or deletion of one or more nucleotides in a DNA sequence.
- Restriction sites
-
A short sequence of DNA that can be recognized and cleaved by restriction enzymes.
- Stoichiometry
-
The quantitative relationship between two or more substances forming a compound.
- Tropism
-
The ability of different viral strains to infect specific cell types or tissues.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Falabella, M., Minczuk, M., Hanna, M.G. et al. Gene therapy for primary mitochondrial diseases: experimental advances and clinical challenges. Nat Rev Neurol 18, 689–698 (2022). https://doi.org/10.1038/s41582-022-00715-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41582-022-00715-9
This article is cited by
-
Induced pluripotent stem cells: ex vivo models for human diseases due to mitochondrial DNA mutations
Journal of Biomedical Science (2023)
-
Current and prospective strategies for advancing the targeted delivery of CRISPR/Cas system via extracellular vesicles
Journal of Nanobiotechnology (2023)
-
Opportunities for mitochondrial disease gene therapy
Nature Reviews Drug Discovery (2023)
-
Mitochondrial DNA editing with mitoARCUS
Nature Metabolism (2023)