Abstract
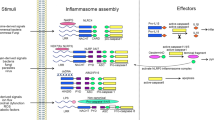
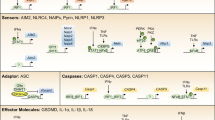
Inflammasomes are supramolecular complexes that form in the cytosol in response to pathogen-associated and damage-associated stimuli, as well as other danger signals that perturb cellular homoeostasis, resulting in host defence responses in the form of cytokine release and programmed cell death (pyroptosis). Inflammasome activity is closely associated with numerous human disorders, including rare genetic syndromes of autoinflammation, cardiovascular diseases, neurodegeneration and cancer. In recent years, a range of inflammasome components and their functions have been discovered, contributing to our knowledge of the overall machinery. Here, we review the latest advances in inflammasome biology from the perspective of structural and mechanistic studies. We focus on the most well-studied components of the canonical inflammasome — NAIP–NLRC4, NLRP3, NLRP1, CARD8 and caspase-1 — as well as caspase-4, caspase-5 and caspase-11 of the noncanonical inflammasome, and the inflammasome effectors GSDMD and NINJ1. These structural studies reveal important insights into how inflammasomes are assembled and regulated, and how they elicit the release of IL-1 family cytokines and induce membrane rupture in pyroptosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lomedico, P. T. et al. Cloning and expression of murine interleukin-1 cDNA in Escherichia coli. Nature 312, 458–462 (1984).
Auron, P. E. et al. Nucleotide sequence of human monocyte interleukin 1 precursor cDNA. Proc. Natl Acad. Sci. USA 81, 7907–7911 (1984).
Van Damme, J. et al. Homogeneous interferon-inducing 22K factor is related to endogenous pyrogen and interleukin-1. Nature 314, 266–268 (1985).
Cerretti, D. P. et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science 256, 97–100 (1992).
Thornberry, N. A. et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356, 768–774 (1992).
Nicholson, D. W. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6, 1028–1042 (1999).
Shalini, S., Dorstyn, L., Dawar, S. & Kumar, S. Old, new and emerging functions of caspases. Cell Death Differ. 22, 526–539 (2015).
Man, S. M. & Kanneganti, T. D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21 (2016).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002). This paper identifies a molecular complex that activates caspase-1, which was termed the inflammasome.
Feldmann, J. et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71, 198–203 (2002).
Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A. & Kolodner, R. D. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301–305 (2001).
Agostini, L. et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 (2004).
Martinon, F., Petrilli, V., Mayor, A., Tardivel, A. & Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006).
So, A., De Smedt, T., Revaz, S. & Tschopp, J. A pilot study of IL-1 inhibition by anakinra in acute gout. Arthritis Res. Ther. 9, R28 (2007).
Hawkins, P. N., Lachmann, H. J., Aganna, E. & McDermott, M. F. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 50, 607–612 (2004).
Jin, T. et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561–571 (2012).
Matyszewski, M. et al. Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome. Nat. Commun. 12, 2735 (2021).
Jin, T., Perry, A., Smith, P., Jiang, J. & Xiao, T. S. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 288, 13225–13235 (2013).
Lu, A. et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 1, 15013 (2015).
Weinert, C., Morger, D., Djekic, A., Grutter, M. G. & Mittl, P. R. Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers. Sci. Rep. 5, 10819 (2015).
Shen, C. et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell 184, 5759–5774 (2021).
Barnett, K. C., Li, S., Liang, K. & Ting, J. P. A 360 degrees view of the inflammasome: mechanisms of activation, cell death, and diseases. Cell 186, 2288–2312 (2023).
Broz, P. & Dixit, V. M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016).
Rathinam, V. A. & Fitzgerald, K. A. Inflammasome complexes: emerging mechanisms and effector functions. Cell 165, 792–800 (2016).
Mangan, M. S. J. et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17, 688 (2018).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Fu, J. & Wu, H. Structural mechanisms of NLRP3 inflammasome assembly and activation. Annu. Rev. Immunol. 41, 301–316 (2023).
Swanson, K. V., Deng, M. & Ting, J. P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Chan, A. H. et al. Caspase-4 dimerisation and D289 auto-processing elicit an interleukin-1beta-converting enzyme. Life Sci. Alliance 6, e202301908 (2023).
Wandel, M. P. et al. Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891 (2020).
Naseer, N. et al. Salmonella enterica serovar Typhimurium induces NAIP/NLRC4- and NLRP3/ASC-independent, caspase-4-dependent inflammasome activation in human intestinal epithelial cells. Infect. Immun. 90, e0066321 (2022).
Knodler, L. A. et al. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe 16, 249–256 (2014).
Xia, S. et al. Gasdermin D pore structure reveals preferential release of mature interleukin-1. Nature 593, 607–611 (2021). This paper reports the structure of the GSDMD pore and illustrates how the charge features in the pore conduit determine the electrostatic filtering that retains pro-IL-1b but releases mature IL-1b.
Liu, X., Xia, S., Zhang, Z., Wu, H. & Lieberman, J. Channelling inflammation: gasdermins in physiology and disease. Nat. Rev. Drug Discov. 20, 384–405 (2021).
Monteleone, M. et al. Interleukin-1 beta maturation triggers its relocation to the plasma membrane for gasdermin-D-dependent and -independent secretion. Cell Rep. 24, 1425–1433 (2018).
Cookson, B. T. & Brennan, M. A. Pro-inflammatory programmed cell death. Trends Microbiol. 9, 113–114 (2001).
Shi, J. et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015).
Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015).
He, W. T. et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 25, 1285–1298 (2015).
Aglietti, R. A. et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl Acad. Sci. USA 113, 7858–7863 (2016).
Ding, J. et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016).
Sborgi, L. et al. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl Acad. Sci. USA 112, 13237–13242 (2015).
Chen, X. et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 26, 1007–1020 (2016).
Russo, H. M. et al. Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J. Immunol. 197, 1353–1367 (2016).
Zheng, D., Liwinski, T. & Elinav, E. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov. 6, 36 (2020).
Christgen, S., Place, D. E. & Kanneganti, T. D. Toward targeting inflammasomes: insights into their regulation and activation. Cell Res. 30, 315–327 (2020).
Tanase, D. M. et al. Portrayal of NLRP3 inflammasome in atherosclerosis: current knowledge and therapeutic targets. Int. J. Mol. Sci. 24, 8162 (2023).
Wang, Y., Sui, Z., Wang, M. & Liu, P. Natural products in attenuating renal inflammation via inhibiting the NLRP3 inflammasome in diabetic kidney disease. Front. Immunol. 14, 1196016 (2023).
Tall, A. R. & Bornfeldt, K. E. Inflammasomes and atherosclerosis: a mixed picture. Circ. Res. 132, 1505–1520 (2023).
Khanmohammadi, S., Ramos-Molina, B. & Kuchay, M. S. NOD-like receptors in the pathogenesis of metabolic (dysfunction)-associated fatty liver disease: therapeutic agents targeting NOD-like receptors. Diabetes Metab. Syndr. 17, 102788 (2023).
Wang, M. et al. Inflammasomes: a rising star on the horizon of COVID-19 pathophysiology. Front. Immunol. 14, 1185233 (2023).
Bauer, S., Hezinger, L., Rexhepi, F., Ramanathan, S. & Kufer, T. A. NOD-like receptors — emerging links to obesity and associated morbidities. Int. J. Mol. Sci. 24, 8595 (2023).
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Chou, W. C., Jha, S., Linhoff, M. W. & Ting, J. P. The NLR gene family: from discovery to present day. Nat. Rev. Immunol. 23, 635–654 (2023).
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009).
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J. & Alnemri, E. S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009).
Heilig, R. & Broz, P. Function and mechanism of the pyrin inflammasome. Eur. J. Immunol. 48, 230–238 (2018).
Lu, A. et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014). This paper reports the first PYD filament structure and the nucleated polymerization mechanism for PYD and CARD in inflammasome assembly.
Richards, N. et al. Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J. Biol. Chem. 276, 39320–39329 (2001).
Lu, A., Kabaleeswaran, V., Fu, T., Magupalli, V. G. & Wu, H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J. Mol. Biol. 426, 1420–1427 (2014).
Hochheiser, I. V. et al. Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation. Sci. Adv. 8, eabn7583 (2022).
Shen, C. et al. Molecular mechanism for NLRP6 inflammasome assembly and activation. Proc. Natl Acad. Sci. USA 116, 2052–2057 (2019).
Dick, M. S., Sborgi, L., Ruhl, S., Hiller, S. & Broz, P. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7, 11929 (2016).
de Alba, E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J. Biol. Chem. 284, 32932–32941 (2009).
Lu, A. et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol. 23, 416–425 (2016).
Li, Y. et al. Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl Acad. Sci. USA 115, 10845–10852 (2018).
Stutz, A., Horvath, G. L., Monks, B. G. & Latz, E. ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040, 91–101 (2013).
Masumoto, J. et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 274, 33835–33838 (1999).
Moriya, M. et al. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry 44, 575–583 (2005).
Gong, Q. et al. Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8. Nat. Commun. 12, 188 (2021).
Hollingsworth, L. R. et al. Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun. 12, 189 (2021).
Zhang, L. et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015).
Wang, L., Sharif, H., Vora, S. M., Zheng, Y. & Wu, H. Structures and functions of the inflammasome engine. J. Allergy Clin. Immunol. 147, 2021–2029 (2021).
Vance, R. E. The NAIP/NLRC4 inflammasomes. Curr. Opin. Immunol. 32, 84–89 (2015).
Hu, Z. et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013). This paper shows the structure of an autoinhibited NLRC4 monomer.
Hu, Z. et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404 (2015). Together with Zhang et al. (2015), this paper shows the structure of the active NLRC4 oligomer, revealing a nucleated polymerization mechanism in NLRC4 oligomerization.
Paidimuddala, B. et al. Mechanism of NAIP-NLRC4 inflammasome activation revealed by cryo-EM structure of unliganded NAIP5. Nat. Struct. Mol. Biol. 30, 159–166 (2023). This paper shows the structure of unliganded NAIP5, revealing how NLRC4 can be bound to NAIP5 before flagellin binding and how NLRC4 is activated conformationally upon flagellin binding.
Schmidt, F. I. et al. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 213, 771–790 (2016).
Man, S. M. et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl Acad. Sci. USA 111, 7403–7408 (2014).
Broz, P., von Moltke, J., Jones, J. W., Vance, R. E. & Monack, D. M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010).
Ross, C. et al. Inflammatory caspases: toward a unified model for caspase activation by inflammasomes. Annu. Rev. Immunol. 40, 249–269 (2022).
Boucher, D. et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 215, 827–840 (2018).
Shi, J. et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011).
Zanoni, I. et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 (2016).
Ross, C., Chan, A. H., Von Pein, J., Boucher, D. & Schroder, K. Dimerization and auto-processing induce caspase-11 protease activation within the non-canonical inflammasome. Life Sci. Alliance 1, e201800237 (2018).
Lee, B. L. et al. Caspase-11 auto-proteolysis is crucial for noncanonical inflammasome activation. J. Exp. Med. 215, 2279–2288 (2018).
Devant, P. et al. Structural insights into cytokine cleavage by inflammatory caspase-4. Nature https://doi.org/10.1038/s41586-023-06751-9 (2023).
Shi, X. et al. Recognition and maturation of IL-18 by caspase-4 noncanonical inflammasome. Nature https://doi.org/10.1038/s41586-023-06742-w (2023).
Gaidt, M. M. & Hornung, V. Pore formation by GSDMD is the effector mechanism of pyroptosis. EMBO J. 35, 2167–2169 (2016).
Sollberger, G. et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol. 3, eaar6689 (2018).
Chen, K. W. et al. Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci. Immunol. 3, eaar6676 (2018).
Yang, J., Zhao, Y., Shi, J. & Shao, F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl Acad. Sci. USA 110, 14408–14413 (2013).
Zhao, Y. et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011).
Kofoed, E. M. & Vance, R. E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011).
Miao, E. A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010).
Kortmann, J., Brubaker, S. W. & Monack, D. M. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 195, 815–819 (2015).
Yang, X. et al. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res. 28, 35–47 (2018). This paper reports the structure of NAIP5 in complex with flagellin and a mutant NLRC4 that cannot oligomerize, revealing how NAIP5 recognizes flagellin.
Tenthorey, J. L. et al. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888–893 (2017). This paper reports the structure of NAIP5 in complex with flagellin and NLRC4, revealing how NAIP5 recognizes flagellin.
Sutterwala, F. S. et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity 24, 317–327 (2006).
Franchi, L., Eigenbrod, T. & Nunez, G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 183, 792–796 (2009).
Bauernfeind, F. G. et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 (2009).
McKee, C. M. & Coll, R. C. NLRP3 inflammasome priming: a riddle wrapped in a mystery inside an enigma. J. Leukoc. Biol. 108, 937–952 (2020).
Hara, H. et al. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates Gram-positive pathogen infection. Cell 175, 1651–1664.e14 (2018).
Puren, A. J., Fantuzzi, G. & Dinarello, C. A. Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1beta are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc. Natl Acad. Sci. USA 96, 2256–2261 (1999).
Marshall, J. D. et al. Regulation of human IL-18 mRNA expression. Clin. Immunol. 90, 15–21 (1999).
Juliana, C. et al. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287, 36617–36622 (2012).
Py, B. F., Kim, M. S., Vakifahmetoglu-Norberg, H. & Yuan, J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013).
Xing, Y. et al. Cutting edge: TRAF6 mediates TLR/IL-1R signaling-induced nontranscriptional priming of the NLRP3 inflammasome. J. Immunol. 199, 1561–1566 (2017).
Fernandes-Alnemri, T. et al. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J. Immunol. 191, 3995–3999 (2013).
Lin, K. M. et al. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proc. Natl Acad. Sci. USA 111, 775–780 (2014).
Song, N. et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 68, 185–197.e6 (2017).
Perregaux, D. & Gabel, C. A. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem. 269, 15195–15203 (1994).
Petrilli, V. et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14, 1583–1589 (2007).
Munoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Mariathasan, S. et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006).
Di, A. et al. The TWIK2 potassium efflux channel in macrophages mediates NLRP3 inflammasome-induced inflammation. Immunity 49, 56–65.e4 (2018).
Murakami, T. et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl Acad. Sci. USA 109, 11282–11287 (2012).
Katsnelson, M. A., Rucker, L. G., Russo, H. M. & Dubyak, G. R. K+ efflux agonists induce NLRP3 inflammasome activation independently of Ca2+ signaling. J. Immunol. 194, 3937–3952 (2015).
Hornung, V. et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008).
Dostert, C. et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320, 674–677 (2008).
Duewell, P. et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010).
Rajamaki, K. et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PloS ONE 5, e11765 (2010).
Halle, A. et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 9, 857–865 (2008).
Dostert, C. et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PloS ONE 4, e6510 (2009).
Shimada, K. et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012).
Nakahira, K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011).
Zhong, Z. et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 (2018).
Xian, H. et al. Oxidized DNA fragments exit mitochondria via mPTP- and VDAC-dependent channels to activate NLRP3 inflammasome and interferon signaling. Immunity 55, 1370–1385.e8 (2022).
Gross, C. J. et al. K+ efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity 45, 761–773 (2016).
Sharif, H. et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019). This paper reports the structure of PYD-deleted NLRP3 in complex with NEK7.
He, Y., Zeng, M. Y., Yang, D., Motro, B. & Nunez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016).
Schmid-Burgk, J. L. et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103–109 (2016).
Shi, H. et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016).
Coll, R. C. et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015).
Schmacke, N. A. et al. IKKβ primes inflammasome formation by recruiting NLRP3 to the trans-Golgi network. Immunity 55, 2271–2284.e7 (2022).
Xiao, L., Magupalli, V. G. & Wu, H. Cryo-EM structures of the active NLRP3 inflammasome disc. Nature 613, 595–600 (2023). This paper reports the NLRP3–NEK7–ASC inflammasome disc in active form, showing the ATP-induced conformational changes.
Hochheiser, I. V. et al. Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604, 184–189 (2022). This paper reports the structure of full-length human NLRP3 in a decameric form in complex with the NLRP3 inhibitor CRID3.
Andreeva, L. et al. NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184, 6299–6312.e22 (2021). This paper reports structures of full-length mouse NLRP3 in oligomeric forms dubbed cages (dodecamer to hexadecamer) that hide the effector PYD inside, and implicates a role for cages in dispersion of the trans-Golgi network.
Ohto, U. et al. Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation. Proc. Natl Acad. Sci. USA 119, e2121353119 (2022).
Misawa, T. et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 (2013).
Chen, J. & Chen, Z. J. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76 (2018).
Zhang, Z. et al. Distinct changes in endosomal composition promote NLRP3 inflammasome activation. Nat. Immunol. 24, 30–41 (2023).
Li, X. et al. MARK4 regulates NLRP3 positioning and inflammasome activation through a microtubule-dependent mechanism. Nat. Commun. 8, 15986 (2017).
Magupalli, V. G. et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science 369, eaas8995 (2020).
Tapia-Abellan, A. et al. Sensing low intracellular potassium by NLRP3 results in a stable open structure that promotes inflammasome activation. Sci. Adv. 7, eabf4468 (2021).
Bittner, Z. A. et al. BTK operates a phospho-tyrosine switch to regulate NLRP3 inflammasome activity. J. Exp. Med. 218, e20201656 (2021).
Subramanian, N., Natarajan, K., Clatworthy, M. R., Wang, Z. & Germain, R. N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013).
Park, S. et al. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J. Immunol. 191, 4358–4366 (2013).
Gangopadhyay, A. et al. NLRP3 licenses NLRP11 for inflammasome activation in human macrophages. Nat. Immunol. 23, 892–903 (2022).
He, M. et al. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 31, 580–591.e5 (2020).
Barry, R. et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 9, 3001 (2018).
Song, H. et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat. Commun. 7, 13727 (2016).
Chavarria-Smith, J., Mitchell, P. S., Ho, A. M., Daugherty, M. D. & Vance, R. E. Functional and evolutionary analyses identify proteolysis as a general mechanism for NLRP1 inflammasome activation. PloS Pathog. 12, e1006052 (2016).
Zhong, F. L. et al. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016).
Tschopp, J., Martinon, F. & Burns, K. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4, 95–104 (2003).
Finger, J. N. et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287, 25030–25037 (2012).
Chui, A. J. et al. N-Terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019).
Taabazuing, C. Y., Griswold, A. R. & Bachovchin, D. A. The NLRP1 and CARD8 inflammasomes. Immunol. Rev. 297, 13–25 (2020).
Sandstrom, A. et al. Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019).
D’Osualdo, A. et al. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PloS ONE 6, e27396 (2011).
Mitchell, P. S., Sandstrom, A. & Vance, R. E. The NLRP1 inflammasome: new mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 60, 37–45 (2019).
Levinsohn, J. L. et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PloS Pathog. 8, e1002638 (2012).
Chavarria-Smith, J. & Vance, R. E. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PloS Pathog. 9, e1003452 (2013).
Boyden, E. D. & Dietrich, W. F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240–244 (2006).
Xu, H. et al. The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J. 38, e101996 (2019).
Gai, K. et al. DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis. 10, 587 (2019).
Okondo, M. C. et al. Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol. 25, 262–267.e5 (2018).
Johnson, D. C. et al. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 24, 1151–1156 (2018).
Zhong, F. L. et al. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 293, 18864–18878 (2018).
Sand, J. et al. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 9, 24 (2018).
Fenini, G. et al. Genome editing of human primary keratinocytes by CRISPR/Cas9 reveals an essential role of the NLRP1 inflammasome in UVB sensing. J. Investig. Dermatol. 138, 2644–2652 (2018).
Wu, C. C., Peterson, A., Zinshteyn, B., Regot, S. & Green, R. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182, 404–416.e14 (2020).
Vind, A. C. et al. ZAKα recognizes stalled ribosomes through partially redundant sensor domains. Mol. Cell 78, 700–713.e7 (2020).
Jenster, L. M. et al. P38 kinases mediate NLRP1 inflammasome activation after ribotoxic stress response and virus infection. J. Exp. Med. 220, e20220837 (2023).
Robinson, K. S. et al. ZAKα-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science 377, 328–335 (2022).
Robinson, K. S. et al. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, eaay2002 (2020).
Tsu, B. V. et al. Diverse viral proteases activate the NLRP1 inflammasome. eLife 10, e60609 (2021).
Bauernfried, S., Scherr, M. J., Pichlmair, A., Duderstadt, K. E. & Hornung, V. Human NLRP1 is a sensor for double-stranded RNA. Science 371, eabd0811 (2021).
Ball, D. P. et al. Oxidized thioredoxin-1 restrains the NLRP1 inflammasome. Sci. Immunol. 7, eabm7200 (2022).
Wang, Q. et al. The NLRP1 and CARD8 inflammasomes detect reductive stress. Cell Rep. 42, 111966 (2023).
Orth-He, E. L. et al. Protein folding stress potentiates NLRP1 and CARD8 inflammasome activation. Cell Rep. 42, 111965 (2023).
Huang, M. et al. Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature 592, 773–777 (2021). This paper reports the structure of an inhibitory ternary complex comprising rat NLRP1, its C-terminal domain and the dipeptidase DPP9.
Hollingsworth, L. R. et al. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature 592, 778–783 (2021). This paper reports the structure of an inhibitory ternary complex comprising human NLRP1, its C-terminal domain and the dipeptidase DPP9.
Levandowski, C. B. et al. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc. Natl Acad. Sci. USA 110, 2952–2956 (2013).
Jin, Y. et al. NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 356, 1216–1225 (2007).
Grandemange, S. et al. A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann. Rheum. Dis. 76, 1191–1198 (2017).
Ball, D. P. et al. Caspase-1 interdomain linker cleavage is required for pyroptosis. Life Sci. Alliance 3, e202000664 (2020).
Jin, T., Curry, J., Smith, P., Jiang, J. & Xiao, T. S. Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1. Proteins 81, 1266–1270 (2013).
Bagnall, R. D. et al. Novel isoforms of the CARD8 (TUCAN) gene evade a nonsense mutation. Eur. J. Hum. Genet. 16, 619–625 (2008).
Sharif, H. et al. Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Immunity 54, 1392–1404.e10 (2021).
Wang, Q. et al. CARD8 is an inflammasome sensor for HIV-1 protease activity. Science 371, eabe1707 (2021).
Tsu, B. V. et al. Host-specific sensing of coronaviruses and picornaviruses by the CARD8 inflammasome. PloS Biol. 21, e3002144 (2023).
Nadkarni, R. et al. Viral proteases activate the CARD8 inflammasome in the human cardiovascular system. J. Exp. Med. 219, e20212117 (2022).
Agard, N. J., Maltby, D. & Wells, J. A. Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell Proteom. 9, 880–893 (2010).
Sarhan, J. et al. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl Acad. Sci. USA 115, E10888–E10897 (2018).
Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018).
Yu, P. et al. Pyroptosis: mechanisms and diseases. Signal Transduct. Target. Ther. 6, 128 (2021).
Xia, X., Wang, X., Zheng, Y., Jiang, J. & Hu, J. What role does pyroptosis play in microbial infection? J. Cell Physiol. 234, 7885–7892 (2019).
Shi, J., Gao, W. & Shao, F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 42, 245–254 (2017).
Kayagaki, N. et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136 (2021).
Liu, Z. et al. Crystal structures of the full-length murine and human gasdermin D reveal mechanisms of autoinhibition, lipid binding, and oligomerization. Immunity 51, 43–49.e4 (2019).
Yang, J. et al. Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor. Proc. Natl Acad. Sci. USA 115, 6792–6797 (2018).
Wang, K. et al. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell 180, 941–955.e20 (2020). This paper reports the structures of caspase-4 and caspase-11, both in complex with GSDMD C-terminus, revealing the important exosite on the caspases for GSDMD recruitment.
Liu, Z. et al. Caspase-1 engages full-length gasdermin D through two distinct interfaces that mediate caspase recruitment and substrate cleavage. Immunity 53, 106–114.e5 (2020). This paper reports the structure of caspase-1 in complex with full-length GSDMD, revealing a two-site engagement, a caspase exosite interaction with GSDMD C-terminus and the caspase active site interaction with the N-terminal to C-terminal linker of GSDMD.
Ruan, J., Xia, S., Liu, X., Lieberman, J. & Wu, H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018). This paper reports the first gasdermin pore structure, revealing its β-barrel architecture, conformational change, lipid interaction and the existence of a prepore oligomer without membrane insertion.
Mulvihill, E. et al. Mechanism of membrane pore formation by human gasdermin-D. EMBO J. 37, e98321 (2018).
Wang, C. et al. Structural basis for GSDMB pore formation and its targeting by IpaH7.8. Nature 616, 590–597 (2023).
Zhong, X. et al. Structural mechanisms for regulation of GSDMB pore-forming activity. Nature 616, 598–605 (2023).
Evavold, C. L. et al. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity 48, 35–44.e6 (2018).
Xie, W. J., Xia, S., Warshel, A. & Wu, H. Electrostatic influence on IL-1 transport through the GSDMD pore. Proc. Natl Acad. Sci. USA 119, e2120287119 (2022).
Keller, M., Ruegg, A., Werner, S. & Beer, H. D. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 (2008).
Chao, Y. Y. et al. Human TH17 cells engage gasdermin E pores to release IL-1α on NLRP3 inflammasome activation. Nat. Immunol. 24, 295–308 (2023).
Araki, T. & Milbrandt, J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J. Neurosci. 20, 187–195 (2000).
Araki, T. & Milbrandt, J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron 17, 353–361 (1996).
Degen, M. et al. Structural basis of NINJ1-mediated plasma membrane rupture in cell death. Nature 618, 1065–1071 (2023). This paper reports the first structure of a NINJ1 oligomer in a double filament and proposes a NINJ1 filament and pore model for membrane damage.
Sahoo, B., Mou, Z., Liu, W., Dubyak, G. & Dai, X. How NINJ1 mediates plasma membrane rupture and why NINJ2 cannot. Preprint at bioRxiv https://doi.org/10.1101/2023.05.31.543175 (2023). This preprint reports the structures of NINJ1 and NINJ2 oligomers in double filaments and proposes why NINJ1 oligomers, but not NINJ2 oligomers, are active for membrane rupture.
David, L. et al. NINJ1 mediates plasma membrane rupture through formation of nanodisc-like rings. Preprint at bioRxiv https://doi.org/10.1101/2023.06.01.543231 (2023). This preprint reports the structure of a NINJ1 oligomer in a ring form with hydrophobic interior, which suggests a nanodisc model for membrane damage.
Acknowledgements
We apologize for incomplete citations owing to space limitations. The work was supported by the National Institutes of Health grants AI124491, AI139914 and AI177778 to H.W. and by the National Health and Medical Research Council of Australia grants 2009677 and 2009075 to K.S.
Author information
Authors and Affiliations
Contributions
J.F. drafted the article, and all authors edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
H.W. is a co-founder and chair of the Scientific Advisory Board of Ventus Therapeutics. K.S. is a co-inventor on patent applications for NLRP3 inhibitors that have been licensed to Inflazome Ltd, which was acquired by Roche. K.S. served on the Scientific Advisory Board of Inflazome, Ireland, and Quench Bio, USA, and serves on a Scientific Advisory Board for Novartis, Switzerland. J.F. declares no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks Z. Hu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Alarmin
-
Alarmins are proteins, peptides, metabolites or others that are released to the outside of a cell in response to immune activation, as a result of transmembrane pore formation, cell injury and lytic death, or degranulation. These endogenous molecules may have chemotactic and/or immune-activating properties to alert the host for defence.
- Cryopyrin-associated periodic syndromes
-
(CAPS). A set of genetic diseases of differing severity caused by autosomal dominant mutations in NLRP3. Patients with CAPS develop spontaneous inflammation and excessive release of the cytokine IL-1β, and may also suffer from arthralgia, deafness and hives.
- Microtubule-organizing centre
-
(MTOC). A structure in eukaryotic cells from which microtubules are nucleated and emanate, which is often synonymous with the centrosome.
- Neutrophil extracellular traps
-
Web-like networks of extracellular DNA, histones and neutrophil granule proteins extruded by neutrophils to trap and damage pathogens.
- Pyroptosis
-
Programmed lytic cell death mediated by cleavage of gasdermin D by caspase-1, caspase-4, caspase-5 and caspase-11, or by other gasdermin family members, that occurs downstream of inflammasome activation or other insults.
- Ribotoxic stress response
-
Activation of stress-associated kinases such as p38 and JNK1 owing to functional ribosome defects that result in inhibition or partial inhibition of protein translation. Downstream signalling can trigger cell cycle arrest, cell death and cytokine production.
- Type III secretion
-
A bacterial secretion system involving a needle-like protein complex used to deliver effector proteins into the host cell cytosol; often found in pathogenic Gram-negative bacteria.
- Unfolded protein response
-
A stress response resulting from the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum that helps cells to reduce the stress through translation suppression, protein degradation and production of chaperones, or that induces cell death with prolonged stress.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fu, J., Schroder, K. & Wu, H. Mechanistic insights from inflammasome structures. Nat Rev Immunol (2024). https://doi.org/10.1038/s41577-024-00995-w
Accepted:
Published:
DOI: https://doi.org/10.1038/s41577-024-00995-w