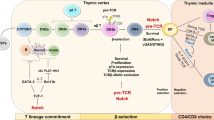
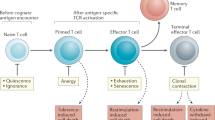
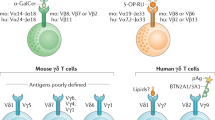
Abstract
The thymus is an evolutionarily conserved organ that supports the development of T cells. Not only does the thymic environment support the rearrangement and expression of diverse T cell receptors but also provides a unique niche for the selection of appropriate T cell clones. Thymic selection ensures that the repertoire of available T cells is both useful (being MHC-restricted) and safe (being self-tolerant). The unique antigen-presentation features of the thymus ensure that the display of self-antigens is optimal to induce tolerance to all types of self-tissue. MHC class-specific functions of CD4+ T helper cells, CD8+ killer T cells and CD4+ regulatory T cells are also established in the thymus. Finally, the thymus provides signals for the development of several minor T cell subsets that promote immune and tissue homeostasis. This Review provides an introductory-level overview of our current understanding of the sophisticated thymic selection mechanisms that ensure T cells are useful and safe.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
01 August 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41577-023-00927-0
References
Burnet, F. M. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J. Clin. 26, 119–121 (1976).
Boehm, T. & Swann, J. B. Origin and evolution of adaptive immunity. Annu. Rev. Anim. Biosci. 2, 259–283 (2014).
Laios, K. The thymus gland in ancient Greek medicine. Horm. Athens Greece 17, 285–286 (2018).
Miller, J. F. Immunological function of the thymus. Lancet 2, 748–749 (1961).
Martinez, C., Kersey, J., Papermaster, B. W. & Good, R. A. Skin homograft survival in thymectomized mice. Proc. Soc. Exp. Biol. Med. 109, 193–196 (1962).
Zinkernagel, R. M. & Doherty, P. C. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248, 701–702 (1974).
Fink, P. J. & Bevan, M. J. H-2 antigens of the thymus determine lymphocyte specificity. J. Exp. Med. 148, 766–775 (1978).
Kisielow, P., Teh, H. S., Blüthmann, H. & von Boehmer, H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature 335, 730–733 (1988).
Kappler, J. W., Staerz, U., White, J. & Marrack, P. C. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature 332, 35–40 (1988).
MacDonald, H. R. et al. T-cell receptor V beta use predicts reactivity and tolerance to Mlsa-encoded antigens. Nature 332, 40–45 (1988).
Kisielow, P., Blüthmann, H., Staerz, U. D., Steinmetz, M. & von Boehmer, H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333, 742–746 (1988).
Anderson, M. S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Lancaster, J. N., Li, Y. & Ehrlich, L. I. R. Chemokine-mediated choreography of thymocyte development and selection. Trends Immunol. 39, 86–98 (2018).
Abramson, J. & Anderson, G. Thymic epithelial cells. Annu. Rev. Immunol. 35, 85–118 (2017). This paper reviews the functions of thymic epithelial cells in positive and negative selection.
Hosokawa, H. & Rothenberg, E. V. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 21, 162–176 (2021). An in-depth review of the transcriptional networks that establish T cell identity early during thymic development.
Zhang, S. L. & Bhandoola, A. Trafficking to the thymus. Curr. Top. Microbiol. Immunol. 373, 87–111 (2014).
Krangel, M. S. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 21, 133–139 (2009).
Schmolka, N., Wencker, M., Hayday, A. C. & Silva-Santos, B. Epigenetic and transcriptional regulation of γδ T cell differentiation: programming cells for responses in time and space. Semin. Immunol. 27, 19–25 (2015). This paper summarizes how γδ T cells develop in the thymus and how their functional properties are established. It also describes key differences between fetal and adult development.
Nitta, T., Murata, S., Ueno, T., Tanaka, K. & Takahama, Y. Thymic microenvironments for T-cell repertoire formation. Adv. Immunol. 99, 59–94 (2008).
Hu, Z., Lancaster, J. N., Sasiponganan, C. & Ehrlich, L. I. R. CCR4 promotes medullary entry and thymocyte–dendritic cell interactions required for central tolerance. J. Exp. Med. 212, 1947–1965 (2015).
James, K. D., Jenkinson, W. E. & Anderson, G. T‐cell egress from the thymus: should I stay or should I go? J. Leukoc. Biol. 104, 275–284 (2018).
Starr, T. K., Jameson, S. C. & Hogquist, K. A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003).
Zerrahn, J., Held, W. & Raulet, D. H. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 88, 627–636 (1997).
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Alam, S. M. et al. T-cell-receptor affinity and thymocyte positive selection. Nature 381, 616–620 (1996).
Punt, J. A., Osborne, B. A., Takahama, Y., Sharrow, S. O. & Singer, A. Negative selection of CD4+ CD8+ thymocytes by T cell receptor-induced apoptosis requires a costimulatory signal that can be provided by CD28. J. Exp. Med. 179, 709–713 (1994).
Breed, E. R., Watanabe, M. & Hogquist, K. A. Measuring thymic clonal deletion at the population level. J. Immunol. Baltim. Md. 1950 202, 3226–3233 (2019).
Buhlmann, J. E., Elkin, S. K. & Sharpe, A. H. A role for the B7-1/B7-2:CD28/CTLA-4 pathway during negative selection. J. Immunol. 170, 5421–5428 (2003).
Salomon, B. et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+ CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity 12, 431–440 (2000).
Tai, X., Cowan, M., Feigenbaum, L. & Singer, A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 6, 152–162 (2005).
Williams, J. A. et al. Regulation of thymic NKT cell development by the B7-CD28 costimulatory pathway. J. Immunol. 181, 907–917 (2008).
Watanabe, M., Lu, Y., Breen, M. & Hodes, R. J. B7-CD28 co-stimulation modulates central tolerance via thymic clonal deletion and Treg generation through distinct mechanisms. Nat. Commun. 11, 6264 (2020). This paper explores how CD28 costimulation contributes to both life (Treg cell generation) and death (clonal deletion) during thymic selection.
Markowitz, J. S., Auchincloss, H., Grusby, M. J. & Glimcher, L. H. Class II-positive hematopoietic cells cannot mediate positive selection of CD4+ T lymphocytes in class II-deficient mice. Proc. Natl Acad. Sci. USA 90, 2779–2783 (1993).
Lo, D. & Sprent, J. Identity of cells that imprint H–2-restricted T-cell specificity in the thymus. Nature 319, 672–675 (1986).
Pishesha, N., Harmand, T. J. & Ploegh, H. L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764 (2022).
Murata, S. et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007).
Gommeaux, J. et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur. J. Immunol. 39, 956–964 (2009).
Honey, K., Nakagawa, T., Peters, C. & Rudensky, A. Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain. J. Exp. Med. 195, 1349–1358 (2002).
Anderson, K. L., Moore, N. C., McLoughlin, D. E., Jenkinson, E. J. & Owen, J. J. Studies on thymic epithelial cells in vitro. Dev. Comp. Immunol. 22, 367–377 (1998).
Sharma, H. & Moroni, L. Recent advancements in regenerative approaches for thymus rejuvenation. Adv. Sci. 8, 2100543 (2021).
Janeway, C. A. The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu. Rev. Immunol. 10, 645–674 (1992).
Robey, E. A. et al. Thymic selection in CD8 transgenic mice supports an instructive model for commitment to a CD4 or CD8 lineage. Cell 64, 99–107 (1991).
Egawa, T. Regulation of CD4 and CD8 coreceptor expression and CD4 versus CD8 lineage decisions. in Advances in Immunology Vol. 125, 1–40 (Elsevier, 2015).
Shinzawa, M. et al. Reversal of the T cell immune system reveals the molecular basis for T cell lineage fate determination in the thymus. Nat. Immunol. 23, 731–742 (2022). This study used genetic engineering of mice to switch the co-receptor proteins encoded by Cd4 and Cd8 gene loci. These mice produce CD4+ cytotoxic T cells and CD8+ T helper cells, and the report explores why such an immune system fails to provide pathogen protection.
Gapin, L. Development of invariant natural killer T cells. Curr. Opin. Immunol. 39, 68–74 (2016). This paper reviews the development of lipid-specific T cells.
Crosby, C. M. & Kronenberg, M. Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 18, 559–574 (2018). This paper reviews the functions of lipid-specific T cells.
Salou, M., Legoux, F. & Lantz, O. MAIT cell development in mice and humans. Mol. Immunol. 130, 31–36 (2021). This paper reviews the development and functions of MAIT cells.
Ruscher, R. & Hogquist, K. A. Development, ontogeny and maintenance of TCRαβ + CD8αα IEL. Curr. Opin. Immunol. 58, 83–88 (2019). This paper reviews the development and functions of intraepithelial lymphocytes.
Stritesky, G. L., Jameson, S. C. & Hogquist, K. A. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 30, 95–114 (2012).
Irla, M. Instructive cues of thymic T cell selection. Annu. Rev. Immunol. 40, 95–119 (2022).
Bouillet, P. et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 (2002).
Pobezinsky, L. A. et al. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol. 13, 569–578 (2012).
McDonald, B. D., Jabri, B. & Bendelac, A. Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 18, 514–525 (2018).
Savage, P. A., Klawon, D. E. J. & Miller, C. H. Regulatory T cell development. Annu. Rev. Immunol. 38, 421–453 (2020). An in-depth consideration of how Treg cells are selected in the thymus.
Bennett, C. L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001).
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Hsieh, C.-S., Zheng, Y., Liang, Y., Fontenot, J. D. & Rudensky, A. Y. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 7, 401–410 (2006).
Malchow, S. et al. AIRE enforces immune tolerance by directing autoreactive T cells into the regulatory T cell lineage. Immunity 44, 1102–1113 (2016).
Toomer, K. H. & Malek, T. R. Cytokine signaling in the development and homeostasis of regulatory T cells. Cold Spring Harb. Perspect. Biol. 10, a028597 (2018).
Klein, L., Robey, E. A. & Hsieh, C.-S. Central CD4+ T cell tolerance: deletion versus regulatory T cell differentiation. Nat. Rev. Immunol. 19, 7–18 (2019). A comprehensive review of CD4+ T cell tolerance.
Lee, H.-M., Bautista, J. L., Scott-Browne, J., Mohan, J. F. & Hsieh, C.-S. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity 37, 475–486 (2012).
Tai, X. et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity 38, 1116–1128 (2013).
Burchill, M. A. et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121 (2008).
Lio, C.-W. J. & Hsieh, C.-S. A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008).
Malhotra, D. et al. Polyclonal CD4+ T cell tolerance is established by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 17, 187–195 (2016). This study tracked T cells reactive to a model self-antigen, GFP, in various mouse strains that express GFP in different tissues. It revealed that T cells specific for abundant self-antigens tend to be deleted, whereas those that recognize rare self-antigens tend to develop into Treg cells.
Nagamine, K. et al. Positional cloning of the APECED gene. Nat. Genet. 17, 393–398 (1997).
Mathis, D. & Benoist, C. Aire. Annu. Rev. Immunol. 27, 287–312 (2009).
Su, M. A. & Anderson, M. S. Aire expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol. 16, 247–258 (2016).
Takaba, H. et al. Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 (2015).
Michelson, D. A., Hase, K., Kaisho, T., Benoist, C. & Mathis, D. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell 185, 2542–2558.e18 (2022). This study describes the development of mTECs into multiple subtypes that mimic epithelial subtypes in peripheral tissues (such as muscle, skin and neuronal tissue). It shows that lineage-defining transcription factors are expressed in mTECs and facilitate the development of such ‘mimetic’ cells, which promote tolerance to the antigens expressed in different tissues.
Breed, E. R., Lee, S. T. & Hogquist, K. A. Directing T cell fate: how thymic antigen presenting cells coordinate thymocyte selection. Semin. Cell Dev. Biol. 84, 2–10 (2018). An overview of the various types of antigen-presenting cell in the thymus and their roles in selection.
Perera, J., Meng, L., Meng, F. & Huang, H. Autoreactive thymic B cells are efficient antigen-presenting cells of cognate self-antigens for T cell negative selection. Proc. Natl Acad. Sci. USA 110, 17011–17016 (2013).
Lu, F.-T. et al. Thymic B cells promote thymus-derived regulatory T cell development and proliferation. J. Autoimmun. 61, 62–72 (2015).
Walters, S. N., Webster, K. E., Daley, S. & Grey, S. T. A role for intrathymic B cells in the generation of natural regulatory T cells. J. Immunol. 193, 170–176 (2014).
Martinez, R. J. et al. Type III interferon drives thymic B cell activation and regulatory T cell generation. Proc. Natl Acad. Sci. USA 120, e2220120120 (2023).
Li, J., Park, J., Foss, D. & Goldschneider, I. Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J. Exp. Med. 206, 607–622 (2009).
Guilliams, M. et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016).
Lei, Y. et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J. Exp. Med. 208, 383–394 (2011).
Bonasio, R. et al. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat. Immunol. 7, 1092–1100 (2006).
Vobořil, M. et al. Toll-like receptor signaling in thymic epithelium controls monocyte-derived dendritic cell recruitment and Treg generation. Nat. Commun. 11, 2361 (2020).
Zegarra-Ruiz, D. F. et al. Thymic development of gut-microbiota-specific T cells. Nature 594, 413–417 (2021). This paper reports that bacterial antigens from the gut are presented in the thymus under some conditions. These surprising findings present a new paradigm for T cell development.
Breed, E. R. et al. Type 2 cytokines in the thymus activate Sirpα + dendritic cells to promote clonal deletion. Nat. Immunol. 23, 1042–1051 (2022).
Ferré, E. M. N., Schmitt, M. M. & Lionakis, M. S. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Front. Pediatr. 9, 723532 (2021).
Policheni, A. N. et al. PD-1 cooperates with AIRE-mediated tolerance to prevent lethal autoimmune disease. Proc. Natl Acad. Sci. USA 119, e2120149119 (2022).
Kishimoto, H. & Sprent, J. Negative selection in the thymus includes semimature T cells. J. Exp. Med. 185, 263–271 (1997).
Xing, Y., Wang, X., Jameson, S. C. & Hogquist, K. A. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17, 565–573 (2016).
McCaughtry, T. M., Wilken, M. S. & Hogquist, K. A. Thymic emigration revisited. J. Exp. Med. 204, 2513–2520 (2007).
Boursalian, T. E., Golob, J., Soper, D. M., Cooper, C. J. & Fink, P. J. Continued maturation of thymic emigrants in the periphery. Nat. Immunol. 5, 418–425 (2004).
James, K. D. et al. Endothelial cells act as gatekeepers for LTβR-dependent thymocyte emigration. J. Exp. Med. 215, 2984–2993 (2018).
Agus, D. B., Surh, C. D. & Sprent, J. Reentry of T cells to the adult thymus is restricted to activated T cells. J. Exp. Med. 173, 1039–1046 (1991).
Santamaria, J., Darrigues, J., van Meerwijk, J. P. M. & Romagnoli, P. Antigen-presenting cells and T-lymphocytes homing to the thymus shape T cell development. Immunol. Lett. 204, 9–15 (2018).
Fink, P. J. The biology of recent thymic emigrants. Annu. Rev. Immunol. 31, 31–50 (2013).
Perera, J. & Huang, H. The development and function of thymic B cells. Cell. Mol. Life Sci. 72, 2657–2663 (2015).
Ardouin, L. et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity 45, 305–318 (2016).
Srinivasan, J. et al. Age-related changes in thymic central tolerance. Front. Immunol. 12, 676236 (2021).
Yang, S., Fujikado, N., Kolodin, D., Benoist, C. & Mathis, D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594 (2015).
Lancaster, J. N. et al. Central tolerance is impaired in the middle-aged thymic environment. Aging Cell 21, e13624 (2022).
Zhang, B. et al. Glimpse of natural selection of long-lived T-cell clones in healthy life. Proc. Natl Acad. Sci. USA 113, 9858–9863 (2016).
Kong, Y., Jing, Y. & Bettini, M. Generation of T cell receptor retrogenic mice. Curr. Protoc. Immunol. 125, e76 (2019).
Perry, J. S. A. et al. Transfer of cell-surface antigens by scavenger receptor CD36 promotes thymic regulatory T cell receptor repertoire development and allo-tolerance. Immunity 48, 923–936.e4 (2018).
Perry, J. S. A. et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426 (2014).
Jenkinson, E. J., Franchi, L. L., Kingston, R. & Owen, J. J. Effect of deoxyguanosine on lymphopoiesis in the developing thymus rudiment in vitro: application in the production of chimeric thymus rudiments. Eur. J. Immunol. 12, 583–587 (1982).
Kurd, N. & Robey, E. A. T-cell selection in the thymus: a spatial and temporal perspective. Immunol. Rev. 271, 114–126 (2016).
Pala, F., Notarangelo, L. D. & Bosticardo, M. Inborn errors of immunity associated with defects of thymic development. Pediatr. Allergy Immunol. 33, e13832 (2022).
Padron, G. T. & Hernandez-Trujillo, V. P. Autoimmunity in primary immunodeficiencies (PID). Clin. Rev. Allergy Immunol. https://doi.org/10.1007/s12016-022-08942-0 (2022).
Akiyama, T. et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29, 423–437 (2008).
Fischer, A. Severe combined immunodeficiencies (SCID). Clin. Exp. Immunol. 122, 143–149 (2000).
Villa, A., Santagata, S., Bozzi, F., Imberti, L. & Notarangelo, L. D. Omenn syndrome: a disorder of Rag1 and Rag2 genes. J. Clin. Immunol. 19, 87–97 (1999).
Walkovich, K. & Vander Lugt, M. ZAP70-related combined immunodeficiency. in GeneReviews® (eds Adam, M. P. et al.) (University of Washington, 1993).
Huang, Q. et al. Molecular feature and therapeutic perspectives of immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. J. Genet. Genom. 47, 17–26 (2020).
Sullivan, K. E. Chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Immunol. Rev. 287, 186–201 (2019).
Hsu, P. et al. CHARGE syndrome: a review. J. Paediatr. Child Health 50, 504–511 (2014).
Reith, W. & Mach, B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19, 331–373 (2001).
Yasumizu, Y. et al. Myasthenia gravis-specific aberrant neuromuscular gene expression by medullary thymic epithelial cells in thymoma. Nat. Commun. 13, 4230 (2022).
Cooper, J. D. History of thymectomy for myasthenia gravis. Thorac. Surg. Clin. 29, 151–158 (2019).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks G. Anderson; E. Robey, who co-reviewed with E. Kim; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- αβ T cells
-
T cells that express a T cell receptor (TCR) composed of rearranged TCRα and TCRβ chains. Most αβ T cells are ‘conventional’ CD8+ or CD4+ T cells that recognize peptide antigens presented by highly polymorphic MHC class I or MHC class II molecules, respectively. However, several less abundant subsets of T cells also express αβ TCRs, including regulatory T cells, invariant natural killer T cells, mucosal associated invariant T cells and intraepithelial lymphocytes.
- γδ T cells
-
T cells that express a T cell receptor (TCR) composed of rearranged TCRγ and TCRδ chains. They are less abundant than αβ T cells in mice and humans. They recognize diverse ligands and have physiological roles in homeostasis and tissue protection.
- Agonist selection
-
The process through which T cell receptor signalling in the thymus directs differentiation into a specific T cell lineage. Agonist selection is required for thymocyte differentiation into regulatory T cells, invariant natural killer T cells, mucosal associated invariant T cells and intraepithelial lymphocytes. In addition to T cell receptor signalling, agonist selection often also requires additional cues for completion of lineage specification, such as costimulatory molecules and cytokines.
- CD4+ T helper cell
-
Upon activation in a lymph node, CD4+ T helper cells acquire effector functions associated with providing activating or ‘helper’ signals to other immune cells, such as B cells and macrophages.
- CD8+ killer T cell
-
Upon activation in a lymph node, CD8+ killer T cells acquire the ability to directly kill target cells through T cell receptor-directed release of cytotoxic molecules.
- Clonal deletion
-
One possible outcome of negative selection. Clonal deletion is initiated by T cell receptor interaction with peptide–MHC and results in programmed cell death (apoptosis), thus eliminating or ‘deleting’ a T cell clone expressing that specific T cell receptor from the developing repertoire.
- Immune tolerance
-
The inability to mount immune responses against self-proteins. Importantly, immune tolerance is distinct from immunodeficiency in that the ability to mount immune responses against foreign antigens is preserved.
- Intraepithelial lymphocytes
-
(IELs). T cells that reside in the epithelial layer of mucosal linings, such as the gastrointestinal tract. IELs are composed of both γδ T cells and αβ T cells. A prominent subpopulation of IELs expresses CD8αα homodimers, unlike conventional CD8+ T cells. It is the thymic precursors of CD8αα IELs that we refer to in this article.
- Invariant natural killer T cells
-
(iNKT cells). A small subset of αβ T cells that recognizes lipid antigens presented by the non-polymorphic MHC-like molecule CD1d.
- Lineage commitment
-
The process by which a double-positive thymocyte acquires the characteristics of either the helper (CD4+) lineage or killer (CD8+) lineage and loses the potential to differentiate into the alternative lineage.
- MHC restriction
-
Refers to the fact that a T cell, through its T cell receptor, recognizes a combinatorial ligand of an MHC molecule presenting a foreign peptide and does not directly recognize the foreign peptide alone.
- Mucosal associated invariant T cells
-
(MAIT cells). A small subset of αβ T cells that recognizes bacterial metabolites presented by the non-polymorphic MHC-like molecule MR1.
- Negative selection
-
The process by which the interaction between T cell receptor and peptide–MHC in the thymus triggers the apoptosis of a thymocyte expressing that receptor or its diversion away from the conventional T cell fate.
- Peptide–MHC
-
(pMHC). During infection, MHC molecules can be loaded with ‘foreign’ or microbial peptides derived from the invading pathogen and presented to T cell receptors on T cells.
- Positive selection
-
The process by which the interaction between T cell receptor and peptide–MHC in the thymus promotes the survival and differentiation of a double-positive thymocyte expressing that receptor.
- Regulatory T cell
-
(Treg cell). A CD4+ MHC class II-restricted T cell that is self-reactive and suppresses the activation of other immune cells by interfering with antigen presentation, producing immunosuppressive cytokines and/or sequestering pro-inflammatory cytokines.
- Self-pMHC
-
Peptide–MHC complexes in which the peptide is derived from a self-protein. MHC molecules expressed at the surface of the cell are typically bound to self-peptides in the absence of infection, as empty MHC molecules are not stable.
- Thymic selection
-
The T cell receptor-dependent cell fate events that shape the repertoire of T cells present in an individual.
- Treg cell selection
-
The process by which T cell receptor signalling in the thymus directs differentiation into the regulatory T cell lineage. Treg cell selection requires recognition of self-peptide–MHC class II molecules and IL-2 signalling.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ashby, K.M., Hogquist, K.A. A guide to thymic selection of T cells. Nat Rev Immunol 24, 103–117 (2024). https://doi.org/10.1038/s41577-023-00911-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-023-00911-8
This article is cited by
-
Adaptive immune receptor repertoire analysis
Nature Reviews Methods Primers (2024)
-
The genetic basis of autoimmunity seen through the lens of T cell functional traits
Nature Communications (2024)