Abstract
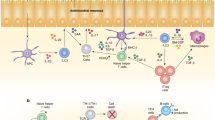
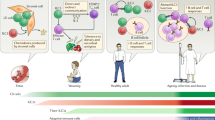
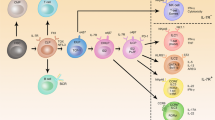
Innate lymphoid cells (ILCs) are important for tissue homeostasis and for the initiation of immune responses. Based on their transcriptional regulation and cytokine profiles, ILCs can be categorized into five subsets with defined phenotypes and functional profiles, but they also have the ability to adapt to local environmental cues by changing these profiles. This plasticity raises the question of the extent to which the cytokine production profiles of ILCs are pre-programmed or are a reflection of the tissue microenvironment. Here, we review recent advances in research on ILCs, with a focus on the plasticity of these cells. We highlight the ability of ILCs to communicate with the surrounding microenvironment and discuss the possible consequences of ILC plasticity for our understanding of the biological roles of these cells. Finally, we discuss how we might use this knowledge of ILC plasticity to develop or improve options for the treatment of inflammatory diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mosmann, T. R., Cherwinski, H., Bond, M. W., Giedlin, M. A. & Coffman, R. L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136, 2348–2357 (1986).
Harrington, L. E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005).
Park, H. et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 (2005).
Duhen, T., Geiger, R., Jarrossay, D., Lanzavecchia, A. & Sallusto, F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10, 857–863 (2009).
Trifari, S., Kaplan, C. D., Tran, E. H., Crellin, N. K. & Spits, H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from TH-17, TH1 and TH2 cells. Nat. Immunol. 10, 864–871 (2009).
Veldhoen, M. et al. Transforming growth factor-β ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9, 1341–1346 (2008).
Lee, Y. J. et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst γδ T, innate lymphoid, and Th cells. J. Immunol. 197, 1460–1470 (2016).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010).
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Price, A. E. et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl Acad. Sci. USA 107, 11489–11494 (2010).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Cupedo, T. et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 10, 66–74 (2009).
Spits, H. & Di Santo, J. P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Spits, H. et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018). This state-of-the-art review summarizes our knowledge of ILCs and updates the ILC classification into five subsets (NK cells, ILC1s, ILC2s, ILC3s and lymphoid tissue inducer cells) based on their development and function.
Spits, H., Bernink, J. H. & Lanier, L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat. Immunol. 17, 758–764 (2016).
Sanos, S. L. et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009).
Luci, C. et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009).
Locksley, R. M. Nine lives: plasticity among T helper cell subsets. J. Exp. Med. 206, 1643–1646 (2009).
DuPage, M. & Bluestone, J. A. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat. Rev. Immunol. 16, 149–163 (2016).
Lim, A. I. & Di Santo, J. P. ILC-poiesis: ensuring tissue ILC differentiation at the right place and time. Eur. J. Immunol. 49, 11–18 (2019).
Cherrier, D. E., Serafini, N. & Di Santo, J. P. Innate lymphoid cell development: a T cell perspective. Immunity 48, 1091–1103 (2018).
Gasteiger, G., Fan, X., Dikiy, S., Lee, S. Y. & Rudensky, A. Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015).
Lim, A. I. et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell 168, 1086–1100.e10 (2017).
Hazenberg, M. D. & Spits, H. Human innate lymphoid cells. Blood 124, 700–709 (2014).
Bar-Ephraim, Y. E. et al. Cross-tissue transcriptomic analysis of human secondary lymphoid organ-residing ILC3s reveals a quiescent state in the absence of inflammation. Cell Rep. 21, 823–833 (2017).
Blom, B. & Spits, H. Development of human lymphoid cells. Annu. Rev. Immunol. 24, 287–320 (2006).
Nagasawa, M. et al. KLRG1 and NKp46 discriminate subpopulations of human CD117+CRTH2– ILCs biased toward ILC2 or ILC3. J. Exp. Med. 216, 1762–1776 (2019). Together with Lim et al. (2017), this paper characterizes circulating ILC precursor populations from human peripheral blood.
Chen, L. et al. CD56 expression marks human group 2 innate lymphoid cell divergence from a shared NK cell and group 3 innate lymphoid cell developmental pathway. Immunity 49, 464–476.e4 (2018).
Thome, J. J. et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med. 22, 72–77 (2016).
Ricardo-Gonzalez, R. R. et al. Tissue-specific pathways extrude activated ILC2 to disseminate type 2 immunity. J. Exp. Med. 217, e20191172 (2020).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13 (2016).
Bjorklund, A. K. et al. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17, 451–460 (2016).
Bar-Ephraim, Y. E. et al. CD62L is a functional and phenotypic marker for circulating innate lymphoid cell precursors. J. Immunol. 202, 171–182 (2019).
Ricardo-Gonzalez, R. R. et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 19, 1093–1099 (2018).
Fang, D. & Zhu, J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 214, 1861–1876 (2017).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity 33, 736–751 (2010).
Cella, M., Otero, K. & Colonna, M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1β reveals intrinsic functional plasticity. Proc. Natl Acad. Sci. USA 107, 10961–10966 (2010).
Crellin, N. K. et al. Regulation of cytokine secretion in human CD127+ LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity 33, 752–764 (2010).
Koues, O. I. et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell 165, 1134–1146 (2016).
Shih, H. Y. et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 165, 1120–1133 (2016).
Stadhouders, R. et al. Epigenome analysis links gene regulatory elements in group 2 innate lymphocytes to asthma susceptibility. J. Allergy Clin. Immunol. 142, 1793–1807 (2018).
Ohne, Y. et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 17, 646–655 (2016).
Xu, W. et al. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep. 10, 2043–2054 (2015).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6–RORγ+ innate lymphoid cells. Nature 494, 261–265 (2013).
Rankin, L. C. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14, 389–395 (2013).
Sciume, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Viant, C. et al. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9, ra46 (2016).
Chea, S. et al. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 9, ra45 (2016).
Mikami, Y. et al. NCR+ ILC3 maintain larger STAT4 reservoir via T-BET to regulate type 1 features upon IL-23 stimulation in mice. Eur. J. Immunol. 48, 1174–1180 (2018).
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015).
Simoni, Y. et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 46, 148–161 (2017).
Bernink, J. H. et al. Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015). This study is one of the first to show that the plasticity of ILCs is associated with the pathology of an inflammatory disease in humans.
Takayama, T. et al. Imbalance of NKp44+NKp46– and NKp44–NKp46+ natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology 139, 882–892 (2010).
Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Li, J. et al. Enrichment of IL-17A+ IFN-γ+ and IL-22+ IFN-γ+ T cell subsets is associated with reduction of NKp44+ ILC3s in the terminal ileum of Crohn’s disease patients. Clin. Exp. Immunol. 190, 143–153 (2017).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013).
Cella, M. et al. Subsets of ILC3–ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019). This paper identifies transitional cell types in the transdifferentiation of human ILC3s to ILC1s in vivo.
Mazzurana, L. et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur. J. Immunol. 49, 1344–1355 (2019).
Parker, M. E. et al. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 217, e20191030 (2020).
Pokrovskii, M. et al. Characterization of transcriptional regulatory networks that promote and restrict identities and functions of intestinal innate lymphoid cells. Immunity 51, 185–197.e6 (2019). Together with Cella et al. (2019), Mazzurana et al. (2019) and Parker et al. (2020), this is the first study of the transcriptional networks that control ILC1 and ILC3 fate and their plasticity.
Silver, J. S. et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 17, 626–635 (2016).
Bal, S. M. et al. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 17, 636–645 (2016).
Lim, A. I. et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 213, 569–583 (2016). Together with Silver et al. (2016) and Bal et al. (2016), this paper shows the involvement of IL-1β and IL-12 in mediating the plasticity of ILC2s to IFNγ-producing ILC1s, which is associated with the pathology of COPD and Crohn’s disease.
Mjosberg, J. et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37, 649–659 (2012).
Golebski, K. et al. IL-1β, IL-23, and TGF-β drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation. Nat. Commun. 10, 2162 (2019).
Bernink, J. H. et al. c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat. Immunol. 20, 992–1003 (2019). Together with Golebski et al. (2019), this paper reports the plasticity of ILC2s to convert into IL-17-producing ILC3-like cells, triggered by cytokines derived from the airway epithelium or dermal stromal cells, in the pathology of human airway or skin inflammation, respectively.
Maier, E., Duschl, A. & Horejs-Hoeck, J. STAT6-dependent and -independent mechanisms in Th2 polarization. Eur. J. Immunol. 42, 2827–2833 (2012).
Zhu, J. Transcriptional regulation of Th2 cell differentiation. Immunol. Cell. Biol. 88, 244–249 (2010).
Stumhofer, J. S. et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8, 1363–1371 (2007).
Hunter, C. A. & Kastelein, R. Interleukin-27: balancing protective and pathological immunity. Immunity 37, 960–969 (2012).
Koenen, H. J. et al. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 112, 2340–2352 (2008).
Tang, Q. et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity 28, 687–697 (2008).
Klatzmann, D. & Abbas, A. K. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat. Rev. Immunol. 15, 283–294 (2015).
Zhang, K. et al. Cutting edge: notch signaling promotes the plasticity of group-2 innate lymphoid cells. J. Immunol. 198, 1798–1803 (2017).
Huang, Y. et al. IL-25-responsive, lineage-negative KLRG1hi cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16, 161–169 (2015).
Huang, Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018). Together with Huang et al. (2015), this paper describes the so-called iILC2s as circulating ILC2s that can produce both IL-17 and IL-13.
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 529, 221–225 (2016).
Martinez-Gonzalez, I. et al. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity 45, 198–208 (2016).
Hochdorfer, T., Winkler, C., Pardali, K. & Mjosberg, J. Expression of c-Kit discriminates between two functionally distinct subsets of human type 2 innate lymphoid cells. Eur. J. Immunol. 49, 884–893 (2019). Together with Bernink et al. (2019), this paper describes two subpopulations of ILC2s in humans.
van de Pavert, S. A. et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014).
Mielke, L. A. et al. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J. Exp. Med. 210, 1117–1124 (2013).
Seehus, C. R. et al. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat. Commun. 8, 1900 (2017).
Morita, H. et al. Induction of human regulatory innate lymphoid cells from group 2 innate lymphoid cells by retinoic acid. J. Allergy Clin. Immunol. 143, 2190–2201.e9 (2019). Together with Seehus et al. (2017), this paper describes the induction of IL-10 production by ILC2s in mice and humans, respectively.
Califano, D. et al. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity 43, 354–368 (2015).
Spooner, C. J. et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 14, 1229–1236 (2013).
Constantinides, M. G., McDonald, B. D., Verhoef, P. A. & Bendelac, A. A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014).
Klose, C. S. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Scoville, S. D. et al. A progenitor cell expressing transcription factor RORγt generates all human innate lymphoid cell subsets. Immunity 44, 1140–1150 (2016).
Renoux, V. M. et al. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity 43, 394–407 (2015).
Xu, W. et al. An Id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity 50, 1054–1068.e3 (2019).
Walker, J. A. et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity 51, 104–118.e7 (2019).
Cortez, V. S. et al. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016).
Cortez, V. S. et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003 (2017).
Gao, Y. et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017).
Cuff, A. O. et al. The obese liver environment mediates conversion of NK cells to a less cytotoxic ILC1-like phenotype. Front. Immunol. 10, 2180 (2019).
Trotta, R. et al. TGF-β utilizes SMAD3 to inhibit CD16-mediated IFN-γ production and antibody-dependent cellular cytotoxicity in human NK cells. J. Immunol. 181, 3784–3792 (2008).
Park, E. et al. Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. eLife 8, e47605 (2019). Together with Cortez et al. (2017), this paper shows the conversion of EOMES+ NK cells to EOMES– ILC1-like cells in tumours and during T. gondii infection and implicates IL-12 and TGFβ in that process.
Takemoto, N., Intlekofer, A. M., Northrup, J. T., Wherry, E. J. & Reiner, S. L. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177, 7515–7519 (2006).
Pikovskaya, O. et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 196, 1449–1454 (2016).
Scoville, S. D., Freud, A. G. & Caligiuri, M. A. Modeling human natural killer cell development in the era of innate lymphoid cells. Front. Immunol. 8, 360 (2017).
Crellin, N. K., Trifari, S., Kaplan, C. D., Cupedo, T. & Spits, H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J. Exp. Med. 207, 281–290 (2010).
Satoh-Takayama, N. et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3–NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207, 273–280 (2010).
Raykova, A. et al. Interleukins 12 and 15 induce cytotoxicity and early NK-cell differentiation in type 3 innate lymphoid cells. Blood Adv. 1, 2679–2691 (2017).
Kotas, M. E. & Locksley, R. M. Why innate lymphoid cells? Immunity 48, 1081–1090 (2018).
Sonnenberg, G. F., Fouser, L. A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Sonnenberg, G. F. et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012).
Qiu, J. & Zhou, L. Aryl hydrocarbon receptor promotes RORγt+ group 3 ILCs and controls intestinal immunity and inflammation. Semin. Immunopathol. 35, 657–670 (2013).
Fragoulis, G. E., Siebert, S. & McInnes, I. B. Therapeutic targeting of IL-17 and IL-23 cytokines in immune-mediated diseases. Annu. Rev. Med. 67, 337–353 (2016).
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007).
Dyring-Andersen, B. et al. Increased number and frequency of group 3 innate lymphoid cells in nonlesional psoriatic skin. Br. J. Dermatol. 170, 609–616 (2014).
Teunissen, M. B. M. et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR+ ILC3 in lesional skin and blood of psoriasis patients. J. Invest. Dermatol. 134, 2351–2360 (2014).
Villanova, F. et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J. Invest. Dermatol. 134, 984–991 (2014).
Everaere, L. et al. Innate lymphoid cells contribute to allergic airway disease exacerbation by obesity. J. Allergy Clin. Immunol. 138, 1309–1318.e11 (2016).
Cai, T. et al. IL-17-producing ST2+ group 2 innate lymphoid cells play a pathogenic role in lung inflammation. J. Allergy Clin. Immunol. 143, 229–244.e9 (2019).
Derycke, L., Zhang, N., Holtappels, G., Dutre, T. & Bachert, C. IL-17A as a regulator of neutrophil survival in nasal polyp disease of patients with and without cystic fibrosis. J. Cyst. Fibros. 11, 193–200 (2012).
Withers, D. R. et al. Transient inhibition of ROR-γt therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat. Med. 22, 319–323 (2016).
Sonnenberg, G. F. & Hepworth, M. R. Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 19, 599–613 (2019).
Zhou, L. et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019).
Hepworth, M. R. et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 498, 113–117 (2013).
Vely, F. et al. Evidence of innate lymphoid cell redundancy in humans. Nat. Immunol. 17, 1291–1299 (2016).
Miller, J. S. & Lanier, L. L. Natural killer cells in cancer immunotherapy. Annu. Rev. Cancer Biol. 3, 77–103 (2019).
Chiossone, L., Dumas, P. Y., Vienne, M. & Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018).
Mattner, J. & Wirtz, S. Friend or foe? The ambiguous role of innate lymphoid cells in cancer development. Trends Immunol. 38, 29–38 (2017).
Kirchberger, S. et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 210, 917–931 (2013).
Nussbaum, K. et al. Tissue microenvironment dictates the fate and tumor-suppressive function of type 3 ILCs. J. Exp. Med. 214, 2331–2347 (2017).
Wagner, M., Moro, K. & Koyasu, S. Plastic heterogeneity of innate lymphoid cells in cancer. Trends Cancer 3, 326–335 (2017).
Xuan, X. et al. ILC3 cells promote the proliferation and invasion of pancreatic cancer cells through IL-22/AKT signaling. Clin. Transl Oncol. https://doi.org/10.1007/s12094-019-02160-5 (2019).
Huber, S. et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263 (2012).
Chan, I. H. et al. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 7, 842–856 (2014).
Koh, J. et al. IL23-producing human lung cancer cells promote tumor growth via conversion of innate lymphoid cell 1 (ILC1) into ILC3. Clin. Cancer Res. 25, 4026–4037 (2019).
Carrega, P. et al. NCR+ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat. Commun. 6, 8280 (2015).
Eisenring, M., vom Berg, J., Kristiansen, G., Saller, E. & Becher, B. IL-12 initiates tumor rejection via lymphoid tissue-inducer cells bearing the natural cytotoxicity receptor NKp46. Nat. Immunol. 11, 1030–1038 (2010).
Trabanelli, S. et al. CD127+ innate lymphoid cells are dysregulated in treatment naive acute myeloid leukemia patients at diagnosis. Haematologica 100, e257–e260 (2015).
Crome, S. Q. et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat. Med. 23, 368–375 (2017).
Blau, H. M. et al. Plasticity of the differentiated state. Science 230, 758–766 (1985).
Blau, H. M. & Baltimore, D. Differentiation requires continuous regulation. J. Cell. Biol. 112, 781–783 (1991).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial–mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Nutt, S. L., Heavey, B., Rolink, A. G. & Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 (1999).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Galli, S. J., Borregaard, N. & Wynn, T. A. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat. Immunol. 12, 1035–1044 (2011).
Szabo, P. A., Miron, M. & Farber, D. L. Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol. 4, eaas9673 (2019).
Eken, A. et al. Fingolimod alters tissue distribution and cytokine production of human and murine innate lymphoid cells. Front. Immunol. 10, 217 (2019).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Weizman, O. E. et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 (2017).
Peng, H. et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123, 1444–1456 (2013).
Roan, F. et al. CD4+ group 1 innate lymphoid cells (ILC) form a functionally distinct ILC subset that is increased in systemic sclerosis. J. Immunol. 196, 2051–2062 (2016).
Acknowledgements
The authors thank I. Martinez-Gonzalez, J. Bernink, B. Heesters, L. Krabbendam and B. Blom for critically reading the manuscript. This work is supported by an advanced grant from the European Research Council to H.S. (No. 341038). S.M.B. is supported by a Lung Foundation Fellowship (No. 6.2.17.196JO).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
H.S. consults for GSK and AIMM Therapeutics and owns stock in AIMM Therapeutics. S.M.B. and K.G. declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thanks A. Freud and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Group 1 ILCs
-
(ILC1s). Group 1 innate lymphoid cells (ILCs) are non-cytotoxic or weakly cytotoxic tissue-resident cells that function as the first line of defence against infections with viruses and certain bacteria. ILC1s produce interferon-γ (IFNγ) and require T-bet for their function. They express CD127 in humans and CD200R in mice, and express NKp46 in both humans and mice.
- Group 2 ILCs
-
(ILC2s). Group 2 innate lymphoid cells (ILCs) mediate parasite expulsion and produce the type 2 cytokines IL-4, IL-5 and IL-13 in response to IL-25, IL-33 and thymic stromal lymphopoietin (TSLP). The transcription factor GATA-binding protein 3 (GATA3) is necessary for ILC2 development and function. Human ILC2s express CRTH2 and high levels of CD161, whereas most mouse ILC2s express ST2.
- Group 3 ILCs
-
(ILC3s). Group 3 innate lymphoid cells (ILCs) are abundant at mucosal sites and are involved in immune responses to extracellular bacteria and the containment of intestinal commensals. ILC3s produce IL-22, by which they promote intestinal homeostasis. Both human and mouse ILC3s can also produce granulocyte–macrophage colony-stimulating factor (GM-CSF). Their development and function depend on retinoic acid receptor-related orphan receptor-γt (RORγt). Two subsets of ILC3s can be distinguished on the basis of cell surface expression of NKp44 in humans and NKp46 in mice.
- Plasticity
-
Plasticity or transdifferentiation is the process by which a cell adapts phenotypically and functionally to a changing local tissue environment.
- RNA velocity analysis
-
Whereas RNA abundance is an indicator of the state of individual cells, RNA velocity is the time derivative of the gene expression state in time. This can be directly estimated by distinguishing between unspliced and spliced mRNAs in common single-cell RNA sequencing protocols. It predicts the future state of individual cells on a timescale of hours.
- Rag2 –/– Il2rg –/– mice
-
A lymphopenic mouse model that lacks T cells, B cells, innate lymphoid cells and natural killer cells.
- Inflammatory ILC2s
-
(iILC2s). These group 2 innate lymphoid cells (ILC2s), which are found predominantly in the circulation, can produce both IL-17 and IL-13, express GATA-binding protein 3 (GATA3) and retinoic acid receptor-related orphan receptor-γt (RORγt), and respond to IL-25. In mice, iILC2s can be identified by the expression of killer cell lectin-like receptor G1 (KLRG1), CC-chemokine receptor 9 (CCR9) and the IL-25 receptor (IL-25R), whereas in humans they lack expression of KLRG1 and CCR9. It should be noted that there is no consensus regarding this terminology in the field and that IL-25-responsive ILC2s can also be found under non-inflammatory conditions.
- Tuft cells
-
Also known as brush cells. ‘Tuft’ refers to the microvilli that project from the surface of these cells. They are secretory epithelial cells found in the intestines and airways that can produce IL-25. They express DCLK1 and TRPM5, and ATOH1 expression is required for their specification.
- Polychromic reporter mice
-
A mouse model consisting of two or more combinatorial reporter genes that enables visualization of more than one protein in the same mouse. Using such mice, researchers have been able to readily identify precursor cells on the basis of expression of one or more transcription factors.
- Common cytokine receptor γ-chain
-
(IL-2Rγ). The common cytokine receptor γ-chain (CD132; encoded by IL2RG) is a component of the receptors for a family of cytokines consisting of IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21.
Rights and permissions
About this article
Cite this article
Bal, S.M., Golebski, K. & Spits, H. Plasticity of innate lymphoid cell subsets. Nat Rev Immunol 20, 552–565 (2020). https://doi.org/10.1038/s41577-020-0282-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-020-0282-9
This article is cited by
-
Sex differences orchestrated by androgens at single-cell resolution
Nature (2024)
-
Group 3 innate lymphoid cells in intestinal health and disease
Nature Reviews Gastroenterology & Hepatology (2024)
-
CD146, a therapeutic target involved in cell plasticity
Science China Life Sciences (2024)
-
Brain endothelial CXCL12 attracts protective natural killer cells during ischemic stroke
Journal of Neuroinflammation (2023)
-
The Notch signaling pathway: a potential target for cancer immunotherapy
Journal of Hematology & Oncology (2023)