Abstract
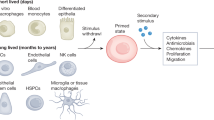
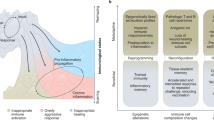
Memories of previous immune events enable barrier tissues to rapidly recall distinct environmental exposures. To effectively inform future responses, these past experiences can be stored in cell types that are long-term residents or essential constituents of tissues. There is an emerging understanding that, in addition to antigen-specific immune cells, diverse haematopoietic, stromal, parenchymal and neuronal cell types can store inflammatory memory. Here, we explore the impact of previous immune activity on various cell lineages with the goal of presenting a unified view of inflammatory memory to environmental exposures (such as allergens, antigens, noxious agents and microorganisms) at barrier tissues. We propose that inflammatory memory is distributed across diverse cell types and stored through shifts in cell states, and we provide a framework to guide future experiments. This distribution and storage may promote adaptation or maladaptation in homeostatic, maintenance and disease settings — especially if the distribution of memory favours cellular cooperation during storage or recall.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Belkaid, Y. & Artis, D. Immunity at the barriers. Eur. J. Immunol. 43, 3096–3097 (2013).
Ordovas-Montanes, J. et al. The regulation of immunological processes by peripheral neurons in homeostasis and disease. Trends Immunol. 36, 578–604 (2015).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Arendt, D. et al. The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016).
Okabe, Y. & Medzhitov, R. Tissue biology perspective on macrophages. Nat. Immunol. 17, 9–17 (2015). This Review discusses the role of macrophages in tissue biology with a focus on cell-type diversification and specialization from an evolutionary and transcriptional perspective.
Kotas, M. E. & Medzhitov, R. Homeostasis, inflammation, and disease susceptibility. Cell 160, 816–827 (2015).
Karin, M. & Clevers, H. Reparative inflammation takes charge of tissue regeneration. Nature 529, 307–315 (2016).
Hedrick, S. M. The acquired immune system: a vantage from beneath. Immunity 21, 607–615 (2004).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
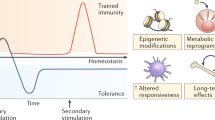
Natoli, G. & Ostuni, R. Adaptation and memory in immune responses. Nat. Immunol. 20, 783–792 (2019). This Review discusses the important concepts of adaptation and memory, providing definitions and molecular properties for these processes.
Schneider, D. & Tate, A. T. Innate immune memory: activation of macrophage killing ability by developmental duties. Curr. Biol. 26, R503–R505 (2016). This Perspective outlines a framework through which to consider memory responses based on the relationship between stimulus levels and response levels.
Netea, M. G. et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016). This Review outlines the properties of trained immunity, and the similarities to and differences from adaptive immunity.
Farber, D. L., Netea, M. G., Radbruch, A., Rajewsky, K. & Zinkernagel, R. M. Immunological memory: lessons from the past and a look to the future. Nat. Rev. Immunol. 16, 124–128 (2016).
Naik, S., Larsen, S. B., Cowley, C. J. & Fuchs, E. Two to tango: dialog between immunity and stem cells in health and disease. Cell 175, 908–920 (2018).
Ordovas-Montanes, J. et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654 (2018). This study demonstrates that human epithelial stem cells may contribute to the persistence of disease by serving as repositories for allergic inflammatory memories.
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017). This study identifies a prolonged memory to acute inflammation that allows murine epidermal stem cells to repair wounds more rapidly on subsequent damage.
Hayday, A., Theodoridis, E., Ramsburg, E. & Shires, J. Intraepithelial lymphocytes: exploring the third way in immunology. Nat. Immunol. 2, 997–1003 (2001).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016).
Satija, R. & Shalek, A. K. Heterogeneity in immune responses: from populations to single cells. Trends Immunol. 35, 219–229 (2014).
Gierahn, T. M. et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017).
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Prakadan, S. M., Shalek, A. K. & Weitz, D. A. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat. Rev. Genet. 18, 345–361 (2017).
Nunez, J. K., Bai, L., Harrington, L. B., Hinder, T. L. & Doudna, J. A. CRISPR immunological memory requires a host factor for specificity. Mol. Cell 62, 824–833 (2016).
Conrath, U., Beckers, G. J., Langenbach, C. J. & Jaskiewicz, M. R. Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119 (2015).
Pradeu, T. & Du Pasquier, L. Immunological memory: what’s in a name? Immunol. Rev. 283, 7–20 (2018).
Ahmed, R. & Gray, D. Immunological memory and protective immunity: understanding their relation. Science 272, 54–60 (1996).
Everitt, A. R. et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 (2012).
Cassone, A. The case for an expanded concept of trained immunity. mBio 9, e00570-18 (2018).
Dai, X. & Medzhitov, R. Inflammation: memory beyond immunity. Nature 550, 460–461 (2017).
Sun, J. C., Ugolini, S. & Vivier, E. Immunological memory within the innate immune system. EMBO J. 33, 1295–1303 (2014).
von Andrian, U. H. & Mackay, C. R. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 343, 1020–1034 (2000).
Steinert, E. M. et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015). This study finds that tissue dissociation techniques underestimate cellular recovery, and quantifies the ratio of parenchymal to tissue-resident CD8 + T cells in selected organs.
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009).
Teijaro, J. R. et al. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 483, 227–231 (2012).
Masopust, D. & Soerens, A. G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 37, 521–546 (2019).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018). This study uses computational predictions and experiments to identify the features of macrophage–fibroblast circuits based on growth factor exchange.
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Crowley, T., Buckley, C. D. & Clark, A. R. Stroma: the forgotten cells of innate immune memory. Clin. Exp. Immunol. 193, 24–36 (2018).
Sohn, C. et al. Prolonged tumor necrosis factor α primes fibroblast-like synoviocytes in a gene-specific manner by altering chromatin. Arthritis Rheumatol. 67, 86–95 (2015).
Wolff, B., Burns, A. R., Middleton, J. & Rot, A. Endothelial cell “memory” of inflammatory stimulation: human venular endothelial cells store interleukin 8 in Weibel-Palade bodies. J. Exp. Med. 188, 1757–1762 (1998).
Foster, S. L., Hargreaves, D. C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Masopust, D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010).
Clark, R. A. et al. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176, 4431–4439 (2006).
Cheroutre, H. & Madakamutil, L. Acquired and natural memory T cells join forces at the mucosal front line. Nat. Rev. Immunol. 4, 290–300 (2004).
Mackay, L. K. & Kallies, A. Transcriptional regulation of tissue-resident lymphocytes. Trends Immunol. 38, 94–103 (2017).
Milner, J. J. & Goldrath, A. W. Transcriptional programming of tissue-resident memory CD8+ T cells. Curr. Opin. Immunol. 51, 162–169 (2018).
Fan, X. & Rudensky, A. Y. Hallmarks of tissue-resident lymphocytes. Cell 164, 1198–1211 (2016).
Iwasaki, A., Foxman, E. F. & Molony, R. D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17, 7–20 (2017).
Radbruch, A. et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 6, 741–750 (2006).
Onodera, T. et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl Acad. Sci. USA 109, 2485–2490 (2012).
Adachi, Y. et al. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 212, 1709–1723 (2015).
Allie, S. R. et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 20, 97–108 (2019).
Oh, J. E. et al. Migrant memory B cells secrete luminal antibody in the vagina. Nature 571, 122–126 (2019).
Landsverk, O. J. et al. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214, 309–317 (2017).
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017). This Perspective provides an evolutionarily-based framework to understand host–microorganism interactions with an emphasis on studying the axes of microbial competition and host control.
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin a for mucosal colonization. Science 360, 795–800 (2018).
Dougan, S. K. et al. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature 503, 406–409 (2013).
Hedrick, S. M. Understanding immunity through the lens of disease ecology. Trends Immunol. 38, 888–903 (2017).
Mora, J. R. et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature 424, 88–93 (2003).
Gerlach, C. et al. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284 (2016).
Mackay, L. K. et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013).
Schenkel, J. M. et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014).
Ariotti, S. et al. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014). Together with Schenkel et al. (2014), this study reports how CD8 + tissue-resident memory T cells activate an alarm function in a tissue, providing protection from an unrelated pathogen.
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019).
Gebhardt, T. et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477, 216–219 (2011).
Roychoudhury, P. et al. Elimination of HSV-2 infected cells is mediated predominantly by paracrine effects of tissue-resident T cell derived cytokines. Preprint at bioRxiv https://doi.org/10.1101/610634 (2019).
Pizzolla, A. et al. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2, eaam6970 (2017).
Khan, T. N., Mooster, J. L., Kilgore, A. M., Osborn, J. F. & Nolz, J. C. Local antigen in nonlymphoid tissue promotes resident memory CD8+ T cell formation during viral infection. J. Exp. Med. 213, 951–966 (2016).
Casey, K. A. et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012).
Mackay, L. K. et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl Acad. Sci. USA 109, 7037–7042 (2012).
Mackay, L. K. et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015).
Mortier, E. et al. Macrophage- and dendritic-cell-derived interleukin-15 receptor α supports homeostasis of distinct CD8+ T cell subsets. Immunity 31, 811–822 (2009).
Mani, V. et al. Migratory DCs activate TGF-β to precondition naive CD8+ T cells for tissue-resident memory fate. Science 366, eaav5728 (2019).
Turner, D. L. et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal. Immunol. 7, 501–510 (2014).
Turner, D. L. et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J. Immunol. 200, 1561–1569 (2018).
Pan, Y. et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017).
Borges da Silva, H. et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature 559, 264–268 (2018).
Stark, R. et al. TRM maintenance is regulated by tissue damage via P2RX7. Sci Immunol. 3, eaau1022 (2018).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Clark, R. A. et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci. Transl Med. 4, 117ra117 (2012).
Collins, N. et al. Skin CD4+ memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun. 7, 11514 (2016).
Oja, A. E. et al. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 11, 654–667 (2018).
Carbone, F. R. & Gebhardt, T. Should I stay or should I go—reconciling clashing perspectives on CD4+ tissue-resident memory T cells. Sci Immunol. 4, eaax5595 (2019).
Klicznik, M. M. et al. Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol. 4, eaav8995 (2019).
Beura, L. K. et al. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216, 1214–1229 (2019).
DiSpirito, J. R. et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol. 3, eaat5861 (2018).
Rosenblum, M. D. et al. Response to self antigen imprints regulatory memory in tissues. Nature 480, 538–542 (2011). This study uses tissue-specific autoantigen expression to identify that regulatory T cells are maintained in tissues and provide enhanced suppression to subsequent autoimmune reactions.
van der Veeken, J. et al. Memory of inflammation in regulatory T cells. Cell 166, 977–990 (2016).
Kadoki, M. et al. Organism-level analysis of vaccination reveals networks of protection across tissues. Cell 171, 398–413.e21 (2017). This study provides a characterization of intra-tissue networks of communication after vaccination and viral infection.
Beura, L. K. et al. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19, 173–182 (2018).
Kotas, M. E. & Locksley, R. M. Why innate lymphoid cells? Immunity 48, 1081–1090 (2018).
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796.e18 (2018).
Mao, K. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Harrison, O. J. et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019).
Sheridan, B. S. et al. γδ T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity 39, 184–195 (2013).
Martinez-Gonzalez, I. et al. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity 45, 198–208 (2016).
Spencer, S. P. et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343, 432–437 (2014). This study illustrates how diet can shift barrier tissues between type 17 and type 2 immunity based on the state of tissue-resident ILCs.
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Quintin, J. et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012).
Monticelli, S. & Natoli, G. Short-term memory of danger signals and environmental stimuli in immune cells. Nat. Immunol. 14, 777–784 (2013).
Ostuni, R. & Natoli, G. Lineages, cell types and functional states: a genomic view. Curr. Opin. Cell Biol. 25, 759–764 (2013).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013). This study identifies latent enhancers in macrophages as regions of the genome in terminally differentiated cells that acquire enhancer-like characteristics after initial stimulation.
Smale, S. T., Tarakhovsky, A. & Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 32, 489–511 (2014).
Yao, Y. et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell 175, 1634–1650.e17 (2018).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Kaufmann, E. et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190.e19 (2018).
Mitroulis, I. et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161.e12 (2018).
Novakovic, B. et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14 (2016).
Daniel, B. et al. The nuclear receptor PPARγ controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity 49, 615–626.e6 (2018).
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Clevers, H., Loh, K. M. & Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014).
Palm, N. W., Rosenstein, R. K. & Medzhitov, R. Allergic host defences. Nature 484, 465–472 (2012).
Beyaz, S. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016). This study illustrates how high-fat diet can alter the intrinsic properties of intestinal epithelial stem and progenitor cells, enhancing stemness and tumorigenic potential.
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320 (2018).
Swamy, M., Jamora, C., Havran, W. & Hayday, A. Epithelial decision makers: in search of the ‘epimmunome’. Nat. Immunol. 11, 656–665 (2010).
Lau, C. M. et al. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 19, 963–972 (2018).
Fanucchi, S. et al. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 51, 138–150 (2019).
Powell, D. W., Pinchuk, I. V., Saada, J. I., Chen, X. & Mifflin, R. C. Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol. 73, 213–237 (2011).
Gieseck, R. L. 3rd, Wilson, M. S. & Wynn, T. A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18, 62–76 (2017).
Stewart, W. E. 2nd, Gosser, L. B. & Lockart, R. Z. Jr Priming: a nonantiviral function of interferon. J. Virol. 7, 792–801 (1971).
Isaacs, A. & Burke, D. C. Mode of action of interferon. Nature 182, 1073–1074 (1958).
Klein, K. et al. The epigenetic architecture at gene promoters determines cell type-specific LPS tolerance. J. Autoimmun. 83, 122–133 (2017).
Crowley, T. et al. Priming in response to pro-inflammatory cytokines is a feature of adult synovial but not dermal fibroblasts. Arthritis Res. Ther. 19, 35 (2017).
Koch, S. R., Lamb, F. S., Hellman, J., Sherwood, E. R. & Stark, R. J. Potentiation and tolerance of Toll-like receptor priming in human endothelial cells. Transl. Res. 180, 53–67.e4 (2017).
Mizoguchi, F. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 9, 789 (2018).
Riol-Blanco, L. et al. Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature 510, 157–161 (2014).
Godinho-Silva, C., Cardoso, F. & Veiga-Fernandes, H. Neuro-immune cell units: a new paradigm in physiology. Annu. Rev. Immunol. 37, 19–46 (2018).
Chavan, S. S., Pavlov, V. A. & Tracey, K. J. Mechanisms and therapeutic relevance of neuro-immune communication. Immunity 46, 927–942 (2017).
Cohen, J. A. et al. Cutaneous TRPV1+ neurons trigger protective innate type 17 anticipatory immunity. Cell 178, 919–932 (2019). This study uses optogenetic activation of heat-sensing sensory neurons to activate an anticipatory type 17 immune response.
Chiu, I. M. et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 501, 52–57 (2013).
Pinho-Ribeiro, F. A. et al. Blocking neuronal signaling to immune cells treats streptococcal invasive infection. Cell 173, 1083–1097.e22 (2018).
Klose, C. S. & Artis, D. Neuronal regulation of innate lymphoid cells. Curr. Opin. Immunol. 56, 94–99 (2018).
Ben-Shaanan, T. L. et al. Activation of the reward system boosts innate and adaptive immunity. Nat. Med. 22, 940–944 (2016).
Choi, G. B. et al. Driving opposing behaviors with ensembles of piriform neurons. Cell 146, 1004–1015 (2011).
Rakoff-Nahoum, S., Foster, K. R. & Comstock, L. E. The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016).
Byers, D. E. et al. Long-term IL-33-producing epithelial progenitor cells in chronic obstructive lung disease. J. Clin. Invest. 123, 3967–3982 (2013).
Lindemans, C. A. et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). This study shows that IL-22 can act directly on intestinal stem cells, illustrating a role for tissue-resident lymphocytes in providing niche signals to epithelial stem cells.
Zenewicz, L. A. et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29, 947–957 (2008).
Huber, S. et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263 (2012).
Scott, H., Solheim, B. G., Brandtzaeg, P. & Thorsby, E. HLA-DR-like antigens in the epithelium of the human small intestine. Scand. J. Immunol. 12, 77–82 (1980).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016). This study identifies tuft cells as key producers of IL-25, and also describes a proto-typical immune cell–epithelial cell circuit in type 2 immunity.
Schneider, C. et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284.e14 (2018).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
Adachi, T. et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 21, 1272–1279 (2015).
Ali, N. et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell 169, 1119–1129.e11 (2017).
Nagao, K. et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 13, 744–752 (2012).
Zaid, A. et al. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl. Acad. Sci. USA 111, 5307–5312 (2014).
Mayassi, T. et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell 176, 967–981.e19 (2019). This study shows how, in coeliac disease, inflammation can deplete innate-like intraepithelial lymphocytes, allowing for accumulation of gluten-reactive cells and an inability to reconstitute the developmentally produced subset.
Park, S. L. et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19, 183–191 (2018).
Dahlgren, M. W. et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50, 707–722 (2019).
Denton, A. E. et al. Type I interferon induces CXCL13 to support ectopic germinal center formation. J. Exp. Med. 216, 621–637 (2019).
Iijima, N. & Iwasaki, A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 346, 93–98 (2014). This study identifies how macrophages and tissue-resident memory CD4 + T cells cooperate to form stable clusters in tissues.
Laidlaw, B. J. et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645 (2014).
Bachem, A. et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297.e5 (2019).
Li, Y. et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Ansaldo, E. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019).
Beura, L. K. et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016).
Yang, Y. et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014).
Omenetti, S. et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity 51, 77–89.e6 (2019).
Rooks, M. G. & Garrett, W. S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016).
Schulthess, J. et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 (2019).
Campbell, C. et al. Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257.e9 (2018).
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Lingwood, D. et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 (2012). This study identifies a ‘pattern-recognition’ function for specific immunoglobulin genes.
Melandri, D. et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 19, 1352–1365 (2018).
Genshaft, A. S. et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 17, 188 (2016).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Shema, E., Bernstein, B. E. & Buenrostro, J. D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 51, 19–25 (2019).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Baron, C. S. & van Oudenaarden, A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 20, 753–765 (2019).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Rubin, A. J. et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176, 361–376.e17 (2019).
Seder, R. A., Darrah, P. A. & Roederer, M. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8, 247–258 (2008).
Martin-Gayo, E. et al. A reproducibility-based computational framework identifies an inducible, enhanced antiviral state in dendritic cells from HIV-1 elite controllers. Genome Biol. 19, 10 (2018).
Hughes, T. K. et al. Highly efficient, massively-parallel single-cell RNA-seq reveals cellular states and molecular features of human skin pathology. Preprint at bioRxiv https://doi.org/10.1101/689273 (2019).
Huang, H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017).
Schleimer, R. P. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu. Rev. Pathol. 12, 331–357 (2017).
Neurath, M. F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 20, 970–979 (2019).
Arijs, I. et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 58, 1612–1619 (2009).
Feagan, B. G. et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 369, 699–710 (2013).
Henikoff, S. & Greally, J. M. Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644–R648 (2016).
Ptashne, M. The chemistry of regulation of genes and other things. J. Biol. Chem. 289, 5417–5435 (2014).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Yosef, N. et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Beyaz, S. et al. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat. Immunol. 18, 184–195 (2017).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes. Dev. 25, 2227–2241 (2011).
Acknowledgements
The authors thank the following individuals for insightful discussions and comments: S. Nyquist, M. Vukovic, S. Kazer, M. Ramseier, V. Miao, C. Kummerlowe, S. Prakadan, E. Christian, A. Hornick, members of the Shalek laboratory, U. von Andrian, C. Borges, Z. Sullivan, V. Mani and S. Naik. This work was supported, in part, by the Damon Runyon Cancer Research Foundation (Howard Hughes Medical Institute Fellow DRG-2274-16; to J.O.-M.), the Richard and Susan Smith Family Foundation (to J.O.-M.), the Searle Scholars Program (to A.K.S.), the Beckman Young Investigator Program (to A.K.S.), the Pew–Stewart Scholars Program for Cancer Research (to A.K.S.), a Sloan Fellowship in Chemistry (to A.K.S.), the US National Institutes of Health (1DP2GM119419, 2U19AI089992, 2R01HL095791, 1U54CA217377, 2P01AI039671, 5U24AI118672, 2RM1HG006193, 1U2CCA23319501, 1R01AI138546, 1R01HL134539 and 1R01DA046277; to A.K.S.), the US Food and Drug Administration (HHSF223201810172C; to A.K.S.) and the Bill and Melinda Gates Foundation (to A.K.S.).
Author information
Authors and Affiliations
Contributions
J.O.-M. contributed to researching the data for the article, discussion of content, and writing and editing the manuscript before submission. A.K.S. contributed to discussion of content, and writing and editing the manuscript before submission. S.B. and S.R.-N. contributed to discussion of content and editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
A.K.S. has received compensation for consulting and SAB membership from Honeycomb Biotechnologies, Cellarity, Cogen Therapeutics and Dahlia Biosciences. All other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thanks A. Iwasaki, G. Natoli, M. Netea and D. Masopust for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Barrier tissues
-
Epithelial tissues that interface directly with the external environment (that is, any surface directly and constantly exposed to the world outside the host), composed of a monolayer, pseudo-stratified or stratified epithelium, as well as an underlying stromal-derived component and other transient, resident or permanent resident cell lineages.
- Immune event
-
Exposure to an environmental stimulus (such as allergens, antigens, noxious agents, diet, pathogens and microbial communities) or a host-derived stimulus (such as metastasis or sterile tissue damage) at a barrier tissue sensed by the host, triggering downstream transcription and/or epigenetic changes in cell state and/or cell composition in the tissue.
- Memory
-
The properties of memory include an altered baseline, sensitivity, rapidity or maximum for a defined response upon secondary challenge to an initiating trigger.
- Adaptive immune memory
-
Classically defined as a memory response by a cell that is considered part of the adaptive immune system (for example, T cells and B cells), based on the ability of its receptor to be formed through the recombination of genetic elements and stably inherited across cell divisions.
- Cell types
-
Developmentally specified cell identity modules that are typically irreversible beyond enforced overexpression of lineage-overriding transcription factors.
- Cell subsets
-
Typically developmentally stable cells, but their programming may be overridden based on niche availability or extreme environments.
- Cell state
-
Characteristics that can be transiently acquired from tissue entry and/or an immune event, are distinct from cellular differentiation and are related to the quality (that is, type of inflammation) of an immune response.
- Gene modules
-
Sets of co-varying genes that may be co-regulated through the activity of one or more transcription factors, or a complex thereof, often associated with a specific cell attribute such as cell type (T cell) or cell state (forkhead box P3 (FOXP3)+ regulatory T cell).
- Inflammatory memory
-
A memory response by any cell lineage to an environmental or host-derived cue, typically acquired during an immune event.
- Protective immunological memory
-
A functionally defensive memory response that enables the host to better respond to secondary challenge after an initial exposure. This function can comprise any of the potential mechanisms that may mediate protective recall, and these same mechanisms may concomitantly or separately mediate immunopathology.
- Innate immune memory
-
Classically defined as a memory response by a cell that is considered part of the innate immune system (for example, macrophages and natural killer cells). However, we favour the use of innate immune memory for memory events triggered by germline-encoded receptors expressed by any cell lineage.
- Lipopolysaccharide (LPS) tolerance
-
Macrophages exposed to sustained stimulation with LPS or high-dose LPS acquire a hypo-responsive state in which sets of inflammatory genes are blunted in their secondary response to LPS or other inflammatory cytokines.
- Tuft cells
-
Rare chemosensory epithelial cells with a ‘tuft-like’ brush of microvilli present in epithelial (primarily mucosal) tissues of mammals, characterized by expression of taste receptors and production of instructive allergic inflammatory cytokines.
- Dendritic epidermal T cells
-
γδ T cell receptor-expressing cells selectively localized in the epidermis that have been identified in rodents and cattle, but not in humans. In mice, essentially all dendritic epidermal T cells express the same T cell receptor constituting a prototypical innate-like T cell.
Rights and permissions
About this article
Cite this article
Ordovas-Montanes, J., Beyaz, S., Rakoff-Nahoum, S. et al. Distribution and storage of inflammatory memory in barrier tissues. Nat Rev Immunol 20, 308–320 (2020). https://doi.org/10.1038/s41577-019-0263-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-019-0263-z
This article is cited by
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization
Cell Death & Differentiation (2024)
-
Cancer cells resistant to immune checkpoint blockade acquire interferon-associated epigenetic memory to sustain T cell dysfunction
Nature Cancer (2023)
-
A subpopulation of lipogenic brown adipocytes drives thermogenic memory
Nature Metabolism (2023)
-
Trained immunity in type 2 immune responses
Mucosal Immunology (2022)
-
Parenteral BCG vaccine induces lung-resident memory macrophages and trained immunity via the gut–lung axis
Nature Immunology (2022)