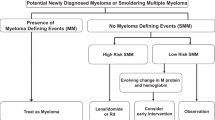
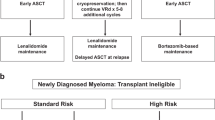
Abstract
Multiple myeloma (MM) is a plasma cell neoplasm that accounts for 2% of all haematological malignancies and predominantly affects older individuals (with a median age at diagnosis of 65–70 years). MM is consistently preceded by the clinically recognized precancerous stages monoclonal gammopathy of undetermined significance and smouldering MM. Thus far, MM has been considered as a single disease entity, but the clinical presentation, response to treatment, and survival outcomes of patients with MM are quite heterogeneous and highly dependent on a set of chromosomal abnormalities that can be identified in nearly all of them. These alterations include primary cytogenetic abnormalities, such as translocations involving chromosome 14q and trisomies of odd-numbered chromosomes, as well as secondary abnormalities, such as deletion of chromosome 17p and amplification of chromosome 1q. The aetiology of myeloma is poorly understood, although different nonoverlapping disease entities can be defined on the basis of their specific primary cytogenetic abnormalities, which have a major role in determining clinical behaviour. This classification might enable the development of better treatment strategies focused on the underlying biology of each specific subtype. Herein, we describe treatment approaches that incorporate the current standard of care for patients with MM along with recommended alterations or improvements that might provide additional clinical benefit for certain subgroups of patients.
Key points
-
The multiple myelomas (MM) comprise a group of highly heterogeneous diseases in terms of underlying genetic abnormalities, clinical presentation, and response to treatment and therefore should not be considered as a single malignancy.
-
The precursor states monoclonal gammopathy of undetermined significance and smouldering MM precede MM and are also characterized by the presence of specific clonal cytogenetic alterations.
-
The presence of primary and secondary abnormalities has been incorporated into several risk-stratification systems for MM developed to predict outcomes and to potentially guide treatment approaches.
-
The initial therapy for patients with MM consists of induction with bortezomib, lenalidomide, and dexamethasone followed by autologous stem cell transplantation in eligible patients and then by maintenance therapy with lenalidomide or bortezomib on the basis of cytogenetic risk factors.
-
At relapse, options for therapy have expanded and include daratumumab, elotuzumab, carfilzomib, ixazomib, and panobinostat.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bhatia, R. Novel approaches to therapy in CML. Hematology Am. Soc. Hematol. Educ. Program 2017, 115–120 (2017).
Tefferi, A. & Barbui, T. Polycythemia vera and essential thrombocythemia: 2017 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 92, 94–108 (2017).
Kapoor, P. et al. Diagnosis and management of waldenstrom macroglobulinemia: mayo stratification of macroglobulinemia and risk-adapted therapy (mSMART) guidelines 2016. JAMA Oncol. 3, 1257–1265 (2017).
Kumar, S. K. et al. Multiple myeloma. Nat. Rev. Dis. Primers 3, 17046 (2017).
Chng, W. J. et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia 28, 269–277 (2014).
Fonseca, R. et al. International myeloma working group molecular classification of multiple myeloma: spotlight review. Leukemia 23, 2210–2221 (2009).
Fonseca, R. Many and multiple myeloma(s). Leukemia 17, 1943–1944 (2003).
Palumbo, A. et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J. Clin. Oncol. 33, 2863–2869 (2015).
Kumar, S. et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood 119, 2100–2105 (2012).
Fonseca, R. et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: an eastern cooperative oncology group study. Cancer Res. 62, 715–720 (2002).
Fonseca, R. et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood 102, 2562–2567 (2003).
Fonseca, R. et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 64, 1546–1558 (2004).
Fonseca, R. et al. Cytogenetic abnormalities in multiple myeloma and related plasma cell disorders: a comparison of conventional cytogenetics to fluorescent in situ hybridization with simultaneous cytoplasmic immunoglobulin staining [abstract]. Blood 90 (Suppl. 1) (1997).
Fonseca, R. Multiple myeloma and FISH (but no CHIPS). Blood 109, 3132–3133 (2007).
Avet-Louseau, H., Daviet, A., Sauner, S., Bataille, R. & Intergroupe Francophone du Myélome. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br. J. Haematol. 111, 1116–1117 (2000).
Avet-Loiseau, H. et al. High incidence of translocations t(11;14) (q13;q32) and t(4;14)(p16;q32) in patients with plasma cell malignancies. Cancer Res. 58, 5640–5645 (1998).
Avet-Loiseau, H. et al. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe francophone du myelome. Cancer Res. 59, 4546–4550 (1999).
Fonseca, R. et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 100, 1417–1424 (2002).
Lopez-Corral, L. et al. Genomic analysis of high-risk smoldering multiple myeloma. Haematologica 97, 1439–1443 (2012).
Rajkumar, S. V. et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia 27, 1738–1744 (2013).
Avet-Loiseau, H. et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J. Clin. Oncol. 28, 4630–4634 (2010).
Jimenez, C. et al. A next-generation sequencing strategy for evaluating the most common genetic abnormalities in multiple myeloma. J. Mol. Diagn. 19, 99–106 (2017).
Kortuem, K. M. et al. Panel sequencing for clinically oriented variant screening and copy number detection in 142 untreated multiple myeloma patients. Blood Cancer J. 6, e397 (2016).
Kortum, K. M. et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M(3) P) in -17p high risk disease. Br. J. Haematol. 168, 507–510 (2015).
Greenberg, A. J. et al. Relationship between initial clinical presentation and the molecular cytogenetic classification of myeloma. Leukemia 28, 398–403 (2014).
Gutierrez, N. C. et al. Correlation between cytogenetic abnormalities and disease characteristics in multiple myeloma: monosomy of chromosome 13 and structural abnormalities of 11q are associated with a high percentage of S-phase plasma cells. Haematologica 85, 1146–1152 (2000).
Kumar, S. et al. High incidence of IgH translocations in monoclonal gammopathies with abnormal free light chain levels. Blood 108, 3514 (2006).
Avet-Loiseau, H. et al. Monosomy 13 is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Intergroupe francophone du myelome. Blood 94, 2583–2589 (1999).
Fonseca, R. et al. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 15, 981–986 (2001).
Binder, M. et al. Prognostic implications of abnormalities of chromosome 13 and the presence of multiple cytogenetic high-risk abnormalities in newly diagnosed multiple myeloma. Blood Cancer J. 7, e600 (2017).
Gertz, M. A. et al. Clinical implications of t(11;14)(q13; q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood 106, 2837–2840 (2005).
Neben, K. et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the international staging system classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica 95, 1150–1157 (2010).
Reece, D. et al. Influence of cytogenetics in patients with relapsed or refractory multiple myeloma treated with lenalidomide plus dexamethasone: adverse effect of deletion 17p13. Blood 114, 522–525 (2009).
Fonseca, R. et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia 20, 2034–2040 (2006).
Hanamura, I. et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 108, 1724–1732 (2006).
Nemec, P. et al. Gain of 1q21 is an unfavorable genetic prognostic factor for multiple myeloma patients treated with high-dose chemotherapy. Biol. Blood Marrow Transplant. 16, 548–554 (2009).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Egan, J. B. et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 120, 1060–1066 (2012).
Walker, B. A. et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 33, 3911–3920 (2015).
Rajkumar, S. V. et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15, e538–e548 (2014).
Mikhael, J. R. et al. Management of newly diagnosed symptomatic multiple myeloma: updated mayo stratification of myeloma and risk-adapted therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 88, 360–376 (2013).
Corre, J. et al. In multiple myeloma, high-risk features are modulated by other chromosomal changes: a large SNParray IFM study. Blood 124, 641 (2014).
Moreau, P. et al. Heterogeneity of t(4;14) in multiple myeloma. Long-term follow-up of 100 cases treated with tandem transplantation in IFM99 trials. Leukemia 21, 2020–2024 (2007).
Neben, K. et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J. Clin. Oncol. 31, 4325–4332 (2013).
Lakshman, A. et al. Natural history of t(11;14) multiple myeloma. Leukemia 32, 131–138 (2017).
Sasaki, K. et al. Impact of t(11;14)(q13; q32) on the outcome of autologous hematopoietic cell transplantation in multiple myeloma. Biol. Blood Marrow Transplant 19, 1227–1232 (2013).
Avet-Loiseau, H. et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood 117, 2009–2011 (2011).
Boersma-Vreugdenhil, G. R. et al. The recurrent translocation t(14;20)(q32; q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br. J. Haematol. 126, 355–363 (2004).
Hebraud, B. et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: the IFM experience. Blood 125, 2095–2100 (2015).
Lode, L. et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 95, 1973–1976 (2010).
Thanendrarajan, S. et al. The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica 102, e364–e367 (2017).
Avet-Loiseau, H. et al. Long-term analysis of the IFM 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J. Clin. Oncol. 30, 1949–1952 (2012).
Fabris, S. et al. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia 21, 1113–1116 (2007).
Sawyer, J. R., Tricot, G., Mattox, S., Jagannath, S. & Barlogie, B. Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 91, 1732–1741 (1998).
Shah, G. L. et al. Gain of chromosome 1q portends worse prognosis in multiple myeloma despite novel agent-based induction regimens and autologous transplantation. Leuk. Lymphoma 58, 1823–1831 (2017).
Greipp, P. R. et al. International staging system for multiple myeloma. J. Clin. Oncol. 23, 3412–3420 (2005).
Kuiper, R. et al. Prediction of high- and low-risk multiple myeloma based on gene expression and the international staging system. Blood 126, 1996–2004 (2015).
van Beers, E. H. et al. Prognostic validation of SKY92 and its combination with ISS in an independent cohort of patients with multiple myeloma. Clin. Lymphoma Myeloma Leuk. 17, 555–562 (2017).
Kuiper, R. et al. A gene expression signature for high-risk multiple myeloma. Leukemia 26, 2406–2413 (2012).
Kumar, S. et al. Gene expression signatures of conventional prognostic factors in multiple. J. Clin. Oncol. 23, 586S (2005).
Kumar, S. K. et al. Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: a multicenter IMWG study. Leukemia 31, 2443–2448 (2017).
Tandon, N. et al. Clinical utility of the revised international staging system in unselected patients with newly diagnosed and relapsed multiple myeloma. Blood Cancer J. 7, e528 (2017).
Majithia, N. et al. Early relapse following initial therapy for multiple myeloma predicts poor outcomes in the era of novel agents. Leukemia 30, 2208–2213 (2016).
Majithia, N. et al. Outcomes of primary refractory multiple myeloma and the impact of novel therapies. Am. J. Hematol. 90, 981–985 (2015).
Kumar, S. K. et al. Early relapse after autologous hematopoietic cell transplantation remains a poor prognostic factor in multiple myeloma but outcomes have improved over time. Leukemia https://doi.org/10.1038/leu.2017.331 (2017).
Kumar, S. Emerging options in multiple myeloma: targeted, immune, and epigenetic therapies. Hematol. Am. Soc. Hematol. Educ. Program 2017, 518–524 (2017).
Shah, J. J. et al. A Phase 1 and 2 study of filanesib alone and in combination with low-dose dexamethasone in relapsed/refractory multiple myeloma. Cancer 123, 4617–4630 (2017).
Kumar, S. K. et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single agent activity in patients with relapsed multiple myeloma. Blood 125, 443–448 (2014).
Kumar, S. et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood 130, 2401–2409 (2017).
Raab, M. S. et al. Phase 1 study update of the novel pan-pim kinase inhibitor LGH447 in patients with relapsed/ refractory multiple myeloma. Blood 124, 301–301 (2014).
Vogl, D. T. et al. Selective inhibition of nuclear export with oral selinexor for treatment of relapsed or refractory multiple myeloma. J. Clin Oncol. 36, 859–866 (2018).
Munshi, N. C. et al. Association of minimal residual disease with superior survival outcomes in patients with multiple myeloma: a meta-analysis. JAMA Oncol. 3, 28–35 (2017).
Kumar, S. et al. International myeloma working group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 17, e328–e346 (2016).
Anderson, K. C. et al. The role of minimal residual disease testing in myeloma treatment selection and drug development: current value and future applications. Clin. Cancer Res. 23, 3980–3993 (2017).
Go, R. S. & Rajkumar, S. V. How I manage monoclonal gammopathy of undetermined significance. Blood 131, 163–173 (2017).
Rajkumar, S. V. MGUS and smoldering multiple myeloma: update on pathogenesis, natural history, and management. Hematology Am. Soc. Hematol. Educ. Program 2005, 340–345 (2005).
Katzmann, J. A. et al. Suppression of uninvolved immunoglobulins defined by heavy/light chain pair suppression is a risk factor for progression of MGUS. Leukemia 27, 208–212 (2013).
Rajkumar, S. V. et al. Presence of monoclonal free light chains in the serum predicts risk of progression in monoclonal gammopathy of undetermined significance (MGUS). Br J Haematol. 127, 308–310 (2004).
Cesana, C. et al. Prognostic factors for malignant transformation in monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. J. Clin. Oncol. 20, 1625–1634 (2002).
Dispenzieri, A. et al. Immunoglobulin free light chain ratio is an independent risk factor for progression of smoldering (asymptomatic) multiple myeloma. Blood 111, 785–789 (2008).
Gonsalves, W. I. et al. Quantification of circulating clonal plasma cells via multiparametric flow cytometry identifies patients with smoldering multiple myeloma at high risk of progression. Leukemia 31, 130–135 (2017).
Kyle, R. A. & Greipp, P. R. Smoldering multiple myeloma. N. Engl. J. Med. 302, 1347–1349 (1980).
Kyle, R. A. et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N. Engl. J. Med. 356, 2582–2590 (2007).
Larsen, J. T. et al. Serum free light chain ratio as a biomarker for high-risk smoldering multiple myeloma. Leukemia 27, 941–946 (2013).
Mateos, M. V. et al. Lenalidomide plus dexamethasone for high-risk smoldering multiple myeloma. N. Engl. J. Med. 369, 438–447 (2013).
Kumar, S. K. et al. Multiple myeloma, version 3.2017, NCCN clinical practice guidelines in oncology. J. Natl Compr. Canc. Netw. 15, 230–269 (2017).
Durie, B. G. et al. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): a randomised, open-label, phase 3 trial. Lancet 389, 519–527 (2017).
Attal, M. et al. Lenalidomide, bortezomib, and dexamethasone with transplantation for myeloma. N. Engl. J. Med. 376, 1311–1320 (2017).
McCarthy, P. L. et al. Lenalidomide maintenance after autologous stem-cell transplantation in newly diagnosed multiple myeloma: a meta-analysis. J. Clin. Oncol. 35, 3279–3289 (2017).
Holstein, S. A. et al. Updated analysis of CALGB (Alliance) 100104 assessing lenalidomide versus placebo maintenance after single autologous stem-cell transplantation for multiple myeloma: a randomised, double-blind, phase 3 trial. Lancet Haematol. 4, e431–e442 (2017).
Laubach, J. et al. Management of relapsed multiple myeloma: recommendations of the international myeloma working group. Leukemia 30, 1005–1017 (2016).
Vu, T. et al. Characteristics of exceptional responders to lenalidomide-based therapy in multiple myeloma. Blood Cancer J. 5, e363 (2015).
Pandey, S. et al. Impact of FISH abnormalities on response to lenalidomide in patients with multiple myeloma. Blood 122, 3210–3210 (2013).
Chretien, M. L. et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood 126, 2713–2719 (2015).
El-Ghammaz, A. M. & Abdelwahed, E. Bortezomib-based induction improves progression-free survival of myeloma patients harboring 17p deletion and/or t(4;14) and overcomes their adverse prognosis. Ann. Hematol. 95, 1315–1321 (2016).
Sonneveld, P. et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: results of the randomized phase III HOVON-65/ GMMG-HD4 trial. J. Clin. Oncol. 30, 2946–2955 (2012).
Cavo, M. et al. Double autologous stem cell transplantation significantly prolongs progression-free survival and overall survival in comparison with single autotransplantation in newly diagnosed multiple myeloma: an analysis of phase 3 EMN02/HO95 study. Blood 130, 401–401 (2017).
Touzeau, C. et al. Deep and sustained response after venetoclax therapy in a patient with very advanced refractory myeloma with translocation t(11;14). Haematologica 102, e112–e114 (2017).
Moreau, P. et al. Promising efficacy and acceptable safety of venetoclax plus bortezomib and dexamethasone in relapsed/refractory MM. Blood 130, 2392–2400 (2017).
Gonsalves, W. I., Buadi, F. K. & Kumar, S. K. Combination therapy incorporating Bcl-2 inhibition with venetoclax for the treatment of refractory primary plasma cell leukemia with t (11;14). Eur. J. Haematol. 100, 215–217 (2018).
Nooka, A. K. et al. Consolidation and maintenance therapy with lenalidomide, bortezomib and dexamethasone (RVD) in high-risk myeloma patients. Leukemia 28, 690–693 (2014).
Kumar, S. et al. Venetoclax monotherapy for relapsed/refractory multiple myeloma: safety and efficacy results from a phase I study. Blood 128, 488 (2016).
Moreau, P. et al. Combination of international scoring system 3, high lactate dehydrogenase, and t(4;14) and/or del(17p) identifies patients with multiple myeloma (MM) treated with front-line autologous stem-cell transplantation at high risk of early MM progression-related death. J. Clin. Oncol. 32, 2173–2180 (2014).
Chin, M. et al. Prevalence and timing of TP53 mutations in del(17p) myeloma and effect on survival. Blood Cancer J. 7, e610 (2017).
Goldschmidt, H. et al. Bortezomib before and after high-dose therapy in myeloma: long-term results from the phase III HOVON-65/GMMG-HD4 trial. Leukemia 32, 383–390 (2017).
Jakubowiak, A. J. et al. A phase 1/2 study of carfilzomib in combination with lenalidomide and low-dose dexamethasone as a frontline treatment for multiple myeloma. Blood 120, 1801–1809 (2012).
Rawstron, A. C. et al. Minimal residual disease assessed by multiparameter flow cytometry in multiple myeloma: impact on outcome in the medical research council myeloma IX study. J. Clin. Oncol. 31, 2540–2547 (2013).
Sawyer, J. R. et al. Evidence of an epigenetic origin for high-risk 1q21 copy number aberrations in multiple myeloma. Blood 125, 3756–3759 (2015).
Sawyer, J. R. et al. Jumping translocations of 1q12 in multiple myeloma: a novel mechanism for deletion of 17p in cytogenetically defined high-risk disease. Blood 123, 2504–2512 (2014).
Jagannath, S. et al. Bortezomib appears to overcome the poor prognosis conferred by chromosome 13 deletion in phase 2 and 3 trials. Leukemia 21, 151–157 (2007).
Xu, J. et al. Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation. Oncogenesis 6, e337 (2017).
Annunziata, C. M. et al. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood 117, 2396–2404 (2011).
Chatterjee, M. et al. Combined disruption of both the MEK/ERK and the IL-6R/STAT3 pathways is required to induce apoptosis of multiple myeloma cells in the presence of bone marrow stromal cells. Blood 104, 3712–3721 (2004).
Rasche, L. et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 8, 268 (2017).
Trudel, S. et al. Biomarker driven phase II clinical trial of trametinib in relapsed/refractory multiple myeloma with sequential addition of the AKT inhibitor, GSK2141795 at time of disease progression to overcome treatment failure: a trial of the princess margaret phase II consortium [poster 4562]. Blood 128, (2016).
Raje, N. et al. Vemurafenib (VEM) in relapsed refractory multiple myeloma harboring BRAF(V600) mutations (V600m): a cohort of the histology-independent ve-basket study. Blood 126, 3 (2015).
Raab, M. S. et al. Spatially divergent clonal evolution in multiple myeloma: overcoming resistance to BRAF inhibition. Blood 127, 2155–2157 (2016).
Decaux, O. et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the intergroupe francophone du myelome. J. Clin. Oncol. 26, 4798–4805 (2008).
Chng, W. J. et al. Gene signature combinations improve prognostic stratification of multiple myeloma patients. Leukemia 30, 1071–1078 (2016).
Shaughnessy, J. D. Jr. et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109, 2276–2284 (2007).
Acknowledgements
S.K.K. and S.V.R. are supported in part by US National Cancer Institute grants CA 107476, CA 168762, and CA186781.
Reviewer information
Nature Reviews Clinical Oncology thanks H. Goldschmidt and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.K.K. and S.V.R. both contributed to researching data for the article, discussions of content, writing the manuscript, and editing and reviewing the manuscript before publication.
Corresponding author
Ethics declarations
Competing interests
S.K.K. has been a non-paid consultant or advisory board member for AbbVie, Celgene, Janssen, Kite Pharma, Merck, and Takeda and is the Principal Investigator in clinical trials supported by Bristol-Myers Squibb, Celgene, Janssen, Kite Pharma, Roche/Genentech, Sanofi, and Takeda. S.V.R. declares no competing interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kumar, S.K., Rajkumar, S.V. The multiple myelomas — current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol 15, 409–421 (2018). https://doi.org/10.1038/s41571-018-0018-y
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-018-0018-y