Abstract
Over the past decade, pharmacogenetic testing has emerged in clinical practice to guide selected cardiovascular therapies. The most common implementation in practice is CYP2C19 genotyping to predict clopidogrel response and assist in selecting antiplatelet therapy after percutaneous coronary intervention. Additional examples include genotyping to guide warfarin dosing and statin prescribing. Increasing evidence exists on outcomes with genotype-guided cardiovascular therapies from multiple randomized controlled trials and observational studies. Pharmacogenetic evidence is accumulating for additional cardiovascular medications. However, data for many of these medications are not yet sufficient to support the use of genotyping for drug prescribing. Ultimately, pharmacogenetics might provide a means to individualize drug regimens for complex diseases such as heart failure, in which the treatment armamentarium includes a growing list of medications shown to reduce morbidity and mortality. However, sophisticated analytical approaches are likely to be necessary to dissect the genetic underpinnings of responses to drug combinations. In this Review, we examine the evidence supporting pharmacogenetic testing in cardiovascular medicine, including that available from several clinical trials. In addition, we describe guidelines that support the use of cardiovascular pharmacogenetics, provide examples of clinical implementation of genotype-guided cardiovascular therapies and discuss opportunities for future growth of the field.
Key points
-
Substantial evidence supports CYP2C19 genotyping to predict the effectiveness of clopidogrel after percutaneous coronary intervention.
-
Data strongly support genetic associations with warfarin dose requirements and risk of simvastatin-induced myopathy, but the use of genotyping in clinical practice to guide prescribing of these drugs is limited.
-
Limited evidence exists to support genetic testing to guide the use of other cardiovascular drugs at present.
-
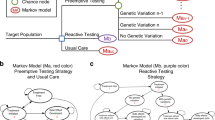
Modern computational approaches can harness large multi-origin data to elucidate genetic predictors of the response to drug treatment for diseases requiring multiple medications targeting various pathogenic pathways.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Roden, D. M. et al. Pharmacogenomics. Lancet 394, 521–532 (2019).
Volpi, S. et al. Research directions in the clinical implementation of pharmacogenomics: an overview of US programs and projects. Clin. Pharmacol. Ther. 103, 778–786 (2018).
Nutescu, E. A. et al. Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy 33, 1156–1164 (2013).
Cavallari, L. H. et al. The IGNITE Pharmacogenetics Working Group: an opportunity for building evidence with pharmacogenetic implementation in a real-world setting. Clin. Transl. Sci. 10, 143–146 (2017).
Cavallari, L. H. et al. Implementation of inpatient models of pharmacogenetics programs. Am. J. Health. Syst. Pharm. 73, 1944–1954 (2016).
Jorgensen, A. L. et al. Implementation of genotype-guided dosing of warfarin with point-of-care genetic testing in three UK clinics: a matched cohort study. BMC Med. 17, 76 (2019).
Relling, M. V. & Klein, T. E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 89, 464–467 (2011).
Ross, C. J. et al. The Canadian Pharmacogenomics Network for Drug Safety: a model for safety pharmacology. Thyroid 20, 681–687 (2010).
Swen, J. J. et al. Pharmacogenetics: from bench to byte — an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011).
Klein, T. E. et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics research network and knowledge base. Pharmacogenomics J. 1, 167–170 (2001).
Food and Drug Administration. Table of Pharmacogenetic Associations. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations (2020).
Collins, K. S. et al. Genotype-guided hydralazine therapy. Am. J. Nephrol. 51, 764–776 (2020).
Thomas, C. D. & Johnson, J. A. Pharmacogenetic factors affecting beta-blocker metabolism and response. Expert Opin. Drug Metab. Toxicol. 16, 953–964 (2020).
Levine, G. N. et al. 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Thorac. Cardiovasc. Surg. 152, 1243–1275 (2016).
Wallentin, L. et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N. Engl. J. Med. 361, 1045–1057 (2009).
Wiviott, S. D. et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N. Engl. J. Med. 357, 2001–2015 (2007).
Dayoub, E. J. et al. Trends in platelet adenosine diphosphate P2Y12 receptor inhibitor use and adherence among antiplatelet-naive patients after percutaneous coronary intervention, 2008–2016. JAMA Intern. Med. 178, 943–950 (2018).
Lombardi, N. et al. Ticagrelor-related late-onset dyspnea as cause of emergency department visit: a 3-year outpatient study. J. Cardiovasc. Med. 19, 284–289 (2018).
Claassens, D. M. F. et al. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. N. Engl. J. Med. 381, 1621–1631 (2019). A major clinical trial of genotype-guided antiplatelet therapy showing similar net adverse clinical outcomes (composite of MACE and major bleeding) but decreased risk of major or minor bleeding with genotype-guided antiplatelet therapy after PCI compared with prasugrel or ticagrelor treatment.
Kazui, M. et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab. Dispos. 38, 92–99 (2010).
Sangkuhl, K., Klein, T. E. & Altman, R. B. Clopidogrel pathway. Pharmacogenet. Genomics 20, 463–465 (2010).
Caudle, K. E. et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 19, 215–223 (2017).
Gaedigk, A. et al. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 103, 399–401 (2018).
Pratt, V. M. et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J. Mol. Diagn. 20, 269–276 (2018).
Scott, S. A. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94, 317–323 (2013).
Mega, J. L. et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 360, 354–362 (2009).
Brandt, J. T. et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J. Thromb. Haemost. 5, 2429–2436 (2007).
Umemura, K., Furuta, T. & Kondo, K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J. Thromb. Haemost. 6, 1439–1441 (2008).
Varenhorst, C. et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur. Heart J. 30, 1744–1752 (2009).
Carreras, E. T. et al. Diabetes mellitus, CYP2C19 genotype, and response to escalating doses of clopidogrel. Insights from the ELEVATE-TIMI 56 trial. Thromb. Haemost. 116, 69–77 (2016).
Mega, J. L. et al. Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA 306, 2221–2228 (2011).
Price, M. J. et al. Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. J. Am. Coll. Cardiol. 59, 1928–1937 (2012).
Mega, J. L. et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation 119, 2553–2560 (2009).
Wallentin, L. et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 376, 1320–1328 (2010).
Sibbing, D. et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 121, 512–518 (2010).
Collet, J. P. et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 373, 309–317 (2009).
Trenk, D. et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J. Am. Coll. Cardiol. 51, 1925–1934 (2008).
Giusti, B. et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am. J. Cardiol. 103, 806–811 (2009).
Mega, J. L. et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304, 1821–1830 (2010).
Combescure, C. et al. Clinical implications of clopidogrel non-response in cardiovascular patients: a systematic review and meta-analysis. J. Thromb. Haemost. 8, 923–933 (2010).
Hulot, J. S. et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J. Am. Coll. Cardiol. 56, 134–143 (2010).
Sorich, M. J., Rowland, A., McKinnon, R. A. & Wiese, M. D. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circ. Cardiovasc. Genet. 7, 895–902 (2014).
Zabalza, M. et al. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart 98, 100–108 (2012).
Bauer, T. et al. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. Br. Med. J. 343, d4588 (2011).
Holmes, M. V., Perel, P., Shah, T., Hingorani, A. D. & Casas, J. P. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA 306, 2704–2714 (2011).
Notarangelo, F. M. et al. Pharmacogenomic approach to selecting antiplatelet therapy in patients with acute coronary syndromes: the PHARMCLO trial. J. Am. Coll. Cardiol. 71, 1869–1877 (2018).
Shen, D. L. et al. Clinical value of CYP2C19 genetic testing for guiding the antiplatelet therapy in a chinese population. J. Cardiovasc. Pharmacol. 67, 232–236 (2016).
Xie, X. et al. Personalized antiplatelet therapy according to CYP2C19 genotype after percutaneous coronary intervention: a randomized control trial. Int. J. Cardiol. 168, 3736–3740 (2013).
Pereira, N. L. et al. Effect of genotype-guided oral P2Y12 inhibitor selection vs conventional clopidogrel therapy on ischemic outcomes after percutaneous coronary intervention: the TAILOR-PCI randomized clinical trial. JAMA 324, 761–771 (2020). A major clinical trial of genotype-guided antiplatelet therapy showing a trend towards reduced risk of MACE and a significant reduction in the risk of multiple recurrent events with genotype-guided therapy after PCI compared with clopidogrel treatment.
Sanchez-Ramos, J. et al. Results of genotype-guided antiplatelet therapy in patients who undergone percutaneous coronary intervention with stent. Int. J. Cardiol. 225, 289–295 (2016).
Holmes, D. R. Jr. et al. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 56, 321–341 (2010).
Empey, P. E. et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin. Pharmacol. Ther. 104, 664–674 (2018).
Angiolillo, D. J. et al. International expert consensus on switching platelet P2Y12 receptor-inhibiting therapies. Circulation 136, 1955–1975 (2017).
Antman, E. M. et al. Early and late benefits of prasugrel in patients with acute coronary syndromes undergoing percutaneous coronary intervention: a TRITON-TIMI 38 (TRial to Assess Improvement in Therapeutic Outcomes by Optimizing Platelet InhibitioN with Prasugrel-Thrombolysis In Myocardial Infarction) analysis. J. Am. Coll. Cardiol. 51, 2028–2033 (2008).
Becker, R. C. et al. Bleeding complications with the P2Y12 receptor antagonists clopidogrel and ticagrelor in the PLATelet inhibition and patient Outcomes (PLATO) trial. Eur. Heart J. 32, 2933–2944 (2011).
Rollini, F., Franchi, F. & Angiolillo, D. J. Switching P2Y12-receptor inhibitors in patients with coronary artery disease. Nat. Rev. Cardiol. 13, 11–27 (2016).
Collet, J. P. et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur. Heart. J. https://doi.org/10.1093/eurheartj/ehaa575 (2020).
Claassens, D. M. & Sibbing, D. De-escalation of antiplatelet treatment in patients with myocardial infarction who underwent percutaneous coronary intervention: a review of the current literature. J. Clin. Med. 9, 2983 (2020).
Peterson, J. F. et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin. Pharmacol. Ther. 100, 67–74 (2016).
Lima, J. J. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin. Pharmacol. Ther. https://doi.org/10.1002/cpt.2015 (2020).
Hicks, J. K. et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015).
Cavallari, L. H. et al. Multisite investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc. Interv. 11, 181–191 (2018). A multicentre collaboration providing CYP2C19 genotyping to guide antiplatelet therapy after PCI as part of clinical care showing an increased risk of MACE in CYP2C19 IMs and PMs treated with clopidogrel compared with other antiplatelet therapy.
Deiman, B. A. et al. Reduced number of cardiovascular events and increased cost-effectiveness by genotype-guided antiplatelet therapy in patients undergoing percutaneous coronary interventions in the Netherlands. Neth. Heart J. 24, 589–599 (2016).
Hulot, J. S. et al. Routine CYP2C19 genotyping to adjust thienopyridine treatment after primary PCI for STEMI: results of the GIANT Study. JACC Cardiovasc. Interv. 13, 621–630 (2020).
Lee, C. R. et al. Impact of the CYP2C19*17 allele on outcomes in patients receiving genotype-guided antiplatelet therapy after percutaneous coronary intervention. Clin. Pharmacol. Ther. 109, 705–715 (2021).
Tiroch, K. A. et al. Protective effect of the CYP2C19*17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am. Heart J. 160, 506–512 (2010).
Gross, L. et al. Genotype-phenotype association and impact on outcomes following guided de-escalation of anti-platelet treatment in acute coronary syndrome patients: the TROPICAL-ACS genotyping substudy. Thromb. Haemost. 118, 1656–1667 (2018).
Wang, Y. et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. JAMA 316, 70–78 (2016).
Pan, Y. et al. Genetic polymorphisms and clopidogrel efficacy for acute ischemic stroke or transient ischemic attack: a systematic review and meta-analysis. Circulation 135, 21–33 (2017).
Diaz-Villamarin, X. et al. Genetic polymorphisms influence on the response to clopidogrel in peripheral artery disease patients following percutaneous transluminal angioplasty. Pharmacogenomics 17, 1327–1338 (2016).
Guo, B. et al. Patients carrying CYP2C19 loss of function alleles have a reduced response to clopidogrel therapy and a greater risk of in-stent restenosis after endovascular treatment of lower extremity peripheral arterial disease. J. Vasc. Surg. 60, 993–1001 (2014).
Hiatt, W. R. et al. Ticagrelor versus clopidogrel in symptomatic peripheral artery disease. N. Engl. J. Med. 376, 32–40 (2017).
Shuldiner, A. R. et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 302, 849–857 (2009).
Mega, J. L. et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet 376, 1312–1319 (2010).
Simon, T. et al. Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 360, 363–375 (2009).
Zhai, Y. et al. Meta-analysis of effects of ABCB1 polymorphisms on clopidogrel response among patients with coronary artery disease. Eur. J. Clin. Pharmacol. 73, 843–854 (2017).
Bouman, H. J. et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat. Med. 17, 110–116 (2011).
Mega, J. L. et al. PON1 Q192R genetic variant and response to clopidogrel and prasugrel: pharmacokinetics, pharmacodynamics, and a meta-analysis of clinical outcomes. J. Thromb. Thrombolysis 41, 374–383 (2016).
Hulot, J. S. et al. CYP2C19 but not PON1 genetic variants influence clopidogrel pharmacokinetics, pharmacodynamics, and clinical efficacy in post-myocardial infarction patients. Circ. Cardiovasc. Interv. 4, 422–428 (2011).
Lewis, J. P. et al. Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. Clin. Pharmacol. Ther. 90, 568–574 (2011).
Lewis, J. P. et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet. Genomics 23, 1–8 (2013).
Lewis, J. P. et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel-treated patients. Eur. Heart J. Cardiovasc. Pharmacother. 6, 203–210 (2020).
Franchi, F., Rollini, F. & Angiolillo, D. J. Defining the link between chronic kidney disease, high platelet reactivity, and clinical outcomes in clopidogrel-treated patients undergoing percutaneous coronary intervention. Circ. Cardiovasc. Interv. 8, e002760 (2015).
Angiolillo, D. J. et al. Impaired responsiveness to the platelet P2Y12 receptor antagonist clopidogrel in patients with type 2 diabetes and coronary artery disease. J. Am. Coll. Cardiol. 64, 1005–1014 (2014).
Angiolillo, D. J. et al. Impact of chronic kidney disease on platelet function profiles in diabetes mellitus patients with coronary artery disease taking dual antiplatelet therapy. J. Am. Coll. Cardiol. 55, 1139–1146 (2010).
Angiolillo, D. J. et al. Derivation, validation, and prognostic utility of a prediction rule for nonresponse to clopidogrel: the ABCD-GENE Score. JACC Cardiovasc. Interv. 13, 606–617 (2020).
Bouziana, S. D. & Tziomalos, K. Clinical relevance of clopidogrel–proton pump inhibitors interaction. World J. Gastrointest. Pharmacol. Ther. 6, 17–21 (2015).
Wadelius, M. et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood 113, 784–792 (2009).
Pirmohamed, M. et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med. 369, 2294–2303 (2013).
Stewart, A., Ganguli, A., FitzGerald, R. & Pirmohamed, M. Variation in warfarin prescribing and dosing in the UK: a national survey of anticoagulation clinics. J. Clin. Pharm. Ther. 40, 466–471 (2015).
Proietti, M., Romanazzi, I., Romiti, G. F., Farcomeni, A. & Lip, G. Y. H. Real-world use of apixaban for stroke prevention in atrial fibrillation: a systematic review and meta-analysis. Stroke 49, 98–106 (2018).
Ntaios, G. et al. Real-world setting comparison of nonvitamin-K antagonist oral anticoagulants versus vitamin-K antagonists for stroke prevention in atrial fibrillation: a systematic review and meta-analysis. Stroke 48, 2494–2503 (2017).
Pare, G. et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation 127, 1404–1412 (2013).
Ziakas, P. D., Kourbeti, I. S., Poulou, L. S., Vlachogeorgos, G. S. & Mylonakis, E. Medicare part D prescribing for direct oral anticoagulants in the United States: cost, use and the “rubber effect”. PLoS ONE 13, e0198674 (2018).
Kjerpeseth, L. J. et al. Trends in use of warfarin and direct oral anticoagulants in atrial fibrillation in Norway, 2010 to 2015. Eur. J. Clin. Pharmacol. 73, 1417–1425 (2017).
Ho, K. H., van Hove, M. & Leng, G. Trends in anticoagulant prescribing: a review of local policies in English primary care. BMC Health Serv. Res. 20, 279 (2020).
Zhu, J., Alexander, G. C., Nazarian, S., Segal, J. B. & Wu, A. W. Trends and variation in oral anticoagulant choice in patients with atrial fibrillation, 2010–2017. Pharmacotherapy 38, 907–920 (2018).
Allabi, A. C. et al. Functional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among Black Africans. Clin. Pharmacol. Ther. 76, 113–118 (2004).
Dickmann, L. J. et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol. Pharmacol. 60, 382–387 (2001).
Liu, Y. et al. Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin. Pharmacol. Ther. 91, 660–665 (2012).
Scordo, M. G. et al. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin. Pharmacol. Ther. 72, 702–710 (2002).
Asiimwe, I. G. et al. Genetic factors influencing warfarin dose in Black-African patients: a systematic review and meta-analysis. Clin. Pharmacol. Ther. 107, 1420–1433 (2020).
Cavallari, L. H. et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin. Pharmacol. Ther. 87, 459–464 (2010).
Higashi, M. K. et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 287, 1690–1698 (2002).
Limdi, N. A. et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin. Pharmacol. Ther. 83, 312–321 (2008).
Rost, S. et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 427, 537–541 (2004).
Wang, D. et al. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 112, 1013–1021 (2008).
Johnson, J. A. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404 (2017).
Limdi, N. A. et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 115, 3827–3834 (2010).
McDonald, M. G., Rieder, M. J., Nakano, M., Hsia, C. K. & Rettie, A. E. CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol. Pharmacol. 75, 1337–1346 (2009).
Danese, E. et al. Effect of CYP4F2, VKORC1, and CYP2C9 in influencing coumarin dose: a single-patient data meta-analysis in more than 15,000 individuals. Clin. Pharmacol. Ther. 105, 1477–1491 (2019).
Bress, A. et al. Effect of NQO1 and CYP4F2 genotypes on warfarin dose requirements in Hispanic-Americans and African-Americans. Pharmacogenomics 13, 1925–1935 (2012).
Cooper, G. M. et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112, 1022–1027 (2008).
Takeuchi, F. et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 5, e1000433 (2009).
Cha, P. C. et al. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum. Mol. Genet. 19, 4735–4744 (2010).
Perera, M. A. et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet 382, 790–796 (2013).
De, T. et al. Association of genetic variants with warfarin-associated bleeding among patients of african descent. JAMA 320, 1670–1677 (2018).
Pisters, R. et al. A novel user-friendly score (HAS-BLED) to assess 1-year risk of major bleeding in patients with atrial fibrillation: the Euro Heart Survey. Chest 138, 1093–1100 (2010).
International Warfarin Pharmacogenetics Consortium. et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 360, 753–764 (2009).
Gage, B. F. et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 84, 326–331 (2008).
Gage, B. F. et al. Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: the GIFT randomized clinical trial. JAMA 318, 1115–1124 (2017). A randomized clinical trial that demonstrated a reduction in the composite of major bleeding, INR ≥4, venous thromboembolism or death in the genotype-guided group compared with the clinically guided dosing group after total joint arthroplasty.
Kimmel, S. E. et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 369, 2283–2293 (2013).
Drozda, K. et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet. Genomics 25, 73–81 (2015).
Arwood, M. J. et al. Anticoagulation endpoints with clinical implementation of warfarin pharmacogenetic dosing in a real-world setting: a proposal for a new pharmacogenetic dosing approach. Clin. Pharmacol. Ther. 101, 675–683 (2017).
Pratt, V. M. et al. Recommendations for clinical warfarin genotyping allele selection: a report of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 22, 847–859 (2020).
U.S. Food & Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labeling. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (2021).
Kaul, K. L., Leonard, D. G., Gonzalez, A. & Garrett, C. T. Oversight of genetic testing: an update. J. Mol. Diagn. 3, 85–91 (2001).
Lee, Y. M. et al. Analysis of comprehensive pharmacogenomic profiling to impact in-hospital prescribing. Pharmacogenet. Genomics 29, 23–30 (2019).
Leary, E., Brilliant, M., Peissig, P. & Griesbach, S. Preliminary outcomes of preemptive warfarin pharmacogenetic testing at a large rural healthcare center. Am. J. Health Syst. Pharm. 76, 387–397 (2019).
Roden, D. M. et al. Benefit of preemptive pharmacogenetic information on clinical outcome. Clin. Pharmacol. Ther. 103, 787–794 (2018).
Marrero, R. J. et al. How to transition from single-gene pharmacogenetic testing to preemptive panel-based testing: a tutorial. Clin. Pharmacol. Ther. 108, 557–565 (2020).
Mega, J. L. et al. Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet 385, 2280–2287 (2015).
Shi, J. et al. Dabigatran etexilate activation is affected by the CES1 genetic polymorphism G143E (rs71647871) and gender. Biochem. Pharmacol. 119, 76–84 (2016).
Buettner, C. et al. Statin use and musculoskeletal pain among adults with and without arthritis. Am. J. Med. 125, 176–182 (2012).
Link, E. et al. SLCO1B1 variants and statin-induced myopathy — a genomewide study. N. Engl. J. Med. 359, 789–799 (2008).
Food and Drug Administration. Drug safety communication: new restrictions, contraindications, and dose limitations for Zocor (simvastatin) to reduce the risk of muscle injury. https://www.fda.gov/drugs/drug-safety-and-availability/fda-drug-safety-communication-new-restrictions-contraindications-and-dose-limitations-zocor (2011).
Hopewell, J. C. et al. Independent risk factors for simvastatin-related myopathy and relevance to different types of muscle symptom. Eur. Heart J. 41, 3336–3342 (2020).
Gupta, A. et al. Adverse events associated with unblinded, but not with blinded, statin therapy in the Anglo-Scandinavian Cardiac Outcomes Trial-Lipid-Lowering Arm (ASCOT-LLA): a randomised double-blind placebo-controlled trial and its non-randomised non-blind extension phase. Lancet 389, 2473–2481 (2017).
Shitara, Y. Clinical importance of OATP1B1 and OATP1B3 in drug–drug interactions. Drug. Metab. Pharmacokinet. 26, 220–227 (2011).
Tirona, R. G., Leake, B. F., Merino, G. & Kim, R. B. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 276, 35669–35675 (2001).
Choi, H. Y. et al. Impact of CYP2D6, CYP3A5, CYP2C19, CYP2A6, SLCO1B1, ABCB1, and ABCG2 gene polymorphisms on the pharmacokinetics of simvastatin and simvastatin acid. Pharmacogenet. Genomics 25, 595–608 (2015).
Pasanen, M. K., Neuvonen, M., Neuvonen, P. J. & Niemi, M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet. Genomics 16, 873–879 (2006).
Ramsey, L. B. et al. The Clinical Pharmacogenetics Implementation Consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin. Pharmacol. Ther. 96, 423–428 (2014).
Tornio, A. et al. SLCO1B1 polymorphism markedly affects the pharmacokinetics of lovastatin acid. Pharmacogenet. Genomics 25, 382–387 (2015).
Voora, D. et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J. Am. Coll. Cardiol. 54, 1609–1616 (2009).
Carr, D. F. et al. SLCO1B1 genetic variant associated with statin-induced myopathy: a proof-of-concept study using the clinical practice research datalink. Clin. Pharmacol. Ther. 94, 695–701 (2013).
Turner, R. M. et al. A genome-wide association study of circulating levels of atorvastatin and its major metabolites. Clin. Pharmacol. Ther. 108, 287–297 (2020). A GWAS showing strong associations between rs4149056 and both circulating atorvastatin levels and muscle-related adverse effects in patients who had been hospitalized with a non-ST-segment elevation ACS and treated with atorvastatin.
Carr, D. F. et al. Genomewide association study of statin-induced myopathy in patients recruited using the UK clinical practice research datalink. Clin. Pharmacol. Ther. 106, 1353–1361 (2019).
Sim, S. C., Edwards, R. J., Boobis, A. R. & Ingelman-Sundberg, M. CYP3A7 protein expression is high in a fraction of adult human livers and partially associated with the CYP3A7*1C allele. Pharmacogenet. Genomics 15, 625–631 (2005).
Becker, M. L. et al. Influence of genetic variation in CYP3A4 and ABCB1 on dose decrease or switching during simvastatin and atorvastatin therapy. Pharmacoepidemiol. Drug Saf. 19, 75–81 (2010).
Fiegenbaum, M. et al. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin. Pharmacol. Ther. 78, 551–558 (2005).
Hoenig, M. R., Walker, P. J., Gurnsey, C., Beadle, K. & Johnson, L. The C3435T polymorphism in ABCB1 influences atorvastatin efficacy and muscle symptoms in a high-risk vascular cohort. J. Clin. Lipidol. 5, 91–96 (2011).
Wilke, R. A., Moore, J. H. & Burmester, J. K. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet. Genomics 15, 415–421 (2005).
Ruano, G. et al. Mechanisms of statin-induced myalgia assessed by physiogenomic associations. Atherosclerosis 218, 451–456 (2011).
Mangravite, L. M. et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 502, 377–380 (2013).
Mega, J. L. et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 385, 2264–2271 (2015).
Natarajan, P. et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 135, 2091–2101 (2017).
Postmus, I. et al. In search for genetic determinants of clinically meaningful differential cardiovascular event reduction by pravastatin in the PHArmacogenetic study of Statins in the Elderly at risk (PHASE)/PROSPER study. Atherosclerosis 235, 58–64 (2014).
Damask, A. et al. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation 141, 624–636 (2020).
Peyser, B. et al. Effects of delivering SLCO1B1 pharmacogenetic information in randomized trial and observational settings. Circ. Genom. Precis. Med. 11, e002228 (2018).
Vassy, J. L. et al. The integrating pharmacogenetics in clinical care (I-PICC) study: protocol for a point-of-care randomized controlled trial of statin pharmacogenetics in primary care. Contemp. Clin. Trials 75, 40–50 (2018).
Danik, J. S. et al. Lack of association between SLCO1B1 polymorphisms and clinical myalgia following rosuvastatin therapy. Am. Heart J. 165, 1008–1014 (2013).
Dutch Pharmacogenetics Working Group, Royal Dutch Pharmacists Association. https://www.knmp.nl/downloads/pharmacogenetic-recommendations-august-2020.pdf (2020).
Lamoureux, F., Duflot, T. & French Network of Pharmacogenetics (RNPGx). Pharmacogenetics in cardiovascular diseases: state of the art and implementation-recommendations of the French National Network of Pharmacogenetics (RNPGx). Therapie 72, 257–267 (2017).
de Keyser, C. E. et al. The SLCO1B1 c.521T>C polymorphism is associated with dose decrease or switching during statin therapy in the Rotterdam study. Pharmacogenet. Genomics 24, 43–51 (2014).
Linskey, D. W. et al. Association of SLCO1B1 c.521T>C (rs4149056) with discontinuation of atorvastatin due to statin-associated muscle symptoms. Pharmacogenet. Genomics 30, 208–211 (2020).
Blake, C. M., Kharasch, E. D., Schwab, M. & Nagele, P. A meta-analysis of CYP2D6 metabolizer phenotype and metoprolol pharmacokinetics. Clin. Pharmacol. Ther. 94, 394–399 (2013).
Baudhuin, L. M. et al. Relation of ADRB1, CYP2D6, and UGT1A1 polymorphisms with dose of, and response to, carvedilol or metoprolol therapy in patients with chronic heart failure. Am. J. Cardiol. 106, 402–408 (2010).
Vieira, C. P., Neves, D. V., Coelho, E. B. & Lanchote, V. L. Effect of CYP2D6 poor metabolizer phenotype on stereoselective nebivolol pharmacokinetics. Drug Metab. Lett. 12, 68–70 (2018).
Lefebvre, J., Poirier, L., Poirier, P., Turgeon, J. & Lacourciere, Y. The influence of CYP2D6 phenotype on the clinical response of nebivolol in patients with essential hypertension. Br. J. Clin. Pharmacol. 63, 575–582 (2007).
Meloche, M. et al. CYP2D6 polymorphism and its impact on the clinical response to metoprolol: a systematic review and meta-analysis. Br. J. Clin. Pharmacol. 86, 1015–1033 (2020).
Zineh, I. et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin. Pharmacol. Ther. 76, 536–544 (2004).
Anstensrud, A. K. et al. Impact of genotype-predicted CYP2D6 metabolism on clinical effects and tolerability of metoprolol in patients after myocardial infarction – a prospective observational study. Eur. J. Clin. Pharmacol. 76, 673–683 (2020).
Batty, J. A. et al. An investigation of CYP2D6 genotype and response to metoprolol CR/XL during dose titration in patients with heart failure: a MERIT-HF substudy. Clin. Pharmacol. Ther. 95, 321–330 (2014).
Johnson, J. A. & Liggett, S. B. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin. Pharmacol. Ther. 89, 366–378 (2011).
Johnson, J. A. et al. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin. Pharmacol. Ther. 74, 44–52 (2003).
Liu, J. et al. β1-Adrenergic receptor polymorphisms influence the response to metoprolol monotherapy in patients with essential hypertension. Clin. Pharmacol. Ther. 80, 23–32 (2006).
Pacanowski, M. A. et al. β-Adrenergic receptor gene polymorphisms and beta-blocker treatment outcomes in hypertension. Clin. Pharmacol. Ther. 84, 715–721 (2008).
Kao, D. P. et al. Effect of bucindolol on heart failure outcomes and heart rate response in patients with reduced ejection fraction heart failure and atrial fibrillation. Eur. J. Heart. Fail. 15, 324–333 (2013).
Liggett, S. B. et al. A polymorphism within a conserved β1-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc. Natl Acad. Sci. USA 103, 11288–11293 (2006).
Cresci, S. et al. Clinical and genetic modifiers of long-term survival in heart failure. J. Am. Coll. Cardiol. 54, 432–444 (2009).
Magvanjav, O. et al. Pharmacogenetic associations of β1-adrenergic receptor polymorphisms with cardiovascular outcomes in the SPS3 trial (Secondary Prevention of Small Subcortical Strokes). Stroke 48, 1337–1343 (2017).
White, H. L. et al. An evaluation of the β-1 adrenergic receptor Arg389Gly polymorphism in individuals with heart failure: a MERIT-HF sub-study. Eur. J. Heart Fail. 5, 463–468 (2003).
Sehrt, D., Meineke, I., Tzvetkov, M., Gultepe, S. & Brockmoller, J. Carvedilol pharmacokinetics and pharmacodynamics in relation to CYP2D6 and ADRB pharmacogenetics. Pharmacogenomics 12, 783–795 (2011).
Huang, J. et al. ADRB2 polymorphism Arg16Gly modifies the natural outcome of heart failure and dictates therapeutic response to beta-blockers in patients with heart failure. Cell Discov. 4, 57 (2018).
Terra, S. G. et al. β-Adrenergic receptor polymorphisms and responses during titration of metoprolol controlled release/extended release in heart failure. Clin. Pharmacol. Ther. 77, 127–137 (2005).
Liggett, S. B. et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat. Med. 14, 510–517 (2008).
Lobmeyer, M. T. et al. Polymorphisms in genes coding for GRK2 and GRK5 and response differences in antihypertensive-treated patients. Pharmacogenet. Genomics 21, 42–49 (2011).
Whelton, P. K. et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 71, e127–e248 (2018).
Gonzalez-Fierro, A. et al. Pharmacokinetics of hydralazine, an antihypertensive and DNA-demethylating agent, using controlled-release formulations designed for use in dosing schedules based on the acetylator phenotype. Int. J. Clin. Pharmacol. Ther. 49, 519–524 (2011).
Han, L. W. et al. Effect of N-acetyltransferase 2 genotype on the pharmacokinetics of hydralazine during pregnancy. J. Clin. Pharmacol. 59, 1678–1689 (2019).
Spinasse, L. B., Santos, A. R., Suffys, P. N., Muxfeldt, E. S. & Salles, G. F. Different phenotypes of the NAT2 gene influences hydralazine antihypertensive response in patients with resistant hypertension. Pharmacogenomics 15, 169–178 (2014).
Schoonen, W. M. et al. Do selected drugs increase the risk of lupus? A matched case–control study. Br. J. Clin. Pharmacol. 70, 588–596 (2010).
Marsden, J. R., Mason, G. G., Coburn, P. R., Rawlins, M. D. & Shuster, S. Drug acetylation and expression of lupus erythematosus. Eur. J. Clin. Pharmacol. 28, 387–390 (1985).
Mazari, L., Ouarzane, M. & Zouali, M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc. Natl Acad. Sci. USA 104, 6317–6322 (2007).
Weber, W. W. & Hein, D. W. N-acetylation pharmacogenetics. Pharmacol. Rev. 37, 25–79 (1985).
Doki, K. et al. Effect of CYP2D6 genotype on flecainide pharmacokinetics in Japanese patients with supraventricular tachyarrhythmia. Eur. J. Clin. Pharmacol. 62, 919–926 (2006).
Doki, K., Sekiguchi, Y., Kuga, K., Aonuma, K. & Homma, M. Serum flecainide S/R ratio reflects the CYP2D6 genotype and changes in CYP2D6 activity. Drug Metab. Pharmacokinet. 30, 257–262 (2015).
Rouini, M. R. & Afshar, M. Effect of CYP2D6 polymorphisms on the pharmacokinetics of propafenone and its two main metabolites. Therapie 72, 373–382 (2017).
Jazwinska-Tarnawska, E. et al. The influence of CYP2D6 polymorphism on the antiarrhythmic efficacy of propafenone in patients with paroxysmal atrial fibrillation during 3 months propafenone prophylactic treatment. Int. J. Clin. Pharmacol. Ther. 39, 288–292 (2001).
Lim, K. S. et al. Changes in the QTc interval after administration of flecainide acetate, with and without coadministered paroxetine, in relation to cytochrome P450 2D6 genotype: data from an open-label, two-period, single-sequence crossover study in healthy Korean male subjects. Clin. Ther. 32, 659–666 (2010).
Arking, D. E. et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet. 46, 826–836 (2014).
Strauss, D. G. et al. Common genetic variant risk score is associated with drug-induced QT prolongation and torsade de pointes risk: a pilot study. Circulation 135, 1300–1310 (2017).
Yancy, C. W. et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation 136, e137–e161 (2017).
Murphy, S. P., Ibrahim, N. E. & Januzzi, J. L. Jr. Heart failure with reduced ejection fraction: a review. JAMA 324, 488–504 (2020).
McNamara, D. M. et al. Endothelial nitric oxide synthase (NOS3) polymorphisms in African Americans with heart failure: results from the A-HeFT trial. J. Card. Fail. 15, 191–198 (2009).
McNamara, D. M. et al. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J. Am. Coll. Cardiol. 44, 2019–2026 (2004).
Segar, M. W. et al. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur. J. Heart Fail. 22, 148–158 (2020).
Shah, S. J. et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 131, 269–279 (2015).
Athreya, A. P. et al. Pharmacogenomics-driven prediction of antidepressant treatment outcomes: a machine-learning approach with multi-trial replication. Clin. Pharmacol. Ther. 106, 855–865 (2019).
Schlotter, F. et al. Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation 138, 377–393 (2018).
Schussler-Fiorenza Rose, S. M. et al. A longitudinal big data approach for precision health. Nat. Med. 25, 792–804 (2019).
Infante, T. et al. Network medicine: a clinical approach for precision medicine and personalized therapy in coronary heart disease. J. Atheroscler. Thromb. 27, 279–302 (2020).
Denny, J. C., Bastarache, L. & Roden, D. M. Phenome-wide association studies as a tool to advance precision medicine. Annu. Rev. Genomics Hum. Genet. 17, 353–373 (2016).
Centers for Medicare & Medicaid Services. Local Coverage Determination (LCD): MolDX: Pharmacogenomics Testing (L38294). https://www.cms.gov/medicare-coverage-database/details/lcd-details.aspx?LCDId=38294&ver=16&Cntrctr=All&UpdatePeriod=889&bc=AAAACAAAAAAA& (2020).
Relling, M. V., Altman, R. B., Goetz, M. P. & Evans, W. E. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 11, 507–509 (2010).
Acknowledgements
J.D.D. and L.H.C. are supported by NIH grant NIH/NHGRI U01 HG00729. L.H.C. is supported by grants NIH/NHLBI R01 HL149752 and NIH/NCATS UL1TR001427.
Author information
Authors and Affiliations
Contributions
J.D.D. and L.H.C. researched the data for the article, provided substantial contributions to discussions of its content, wrote the article and undertook review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cardiology thanks J. W. Jukema and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CPIC: https://cpicpgx.org/
PharmGKB: https://www.pharmgkb.org
Warfarin Dosing: http://www.warfarindosing.org/Source/Home.aspx
Rights and permissions
About this article
Cite this article
Duarte, J.D., Cavallari, L.H. Pharmacogenetics to guide cardiovascular drug therapy. Nat Rev Cardiol 18, 649–665 (2021). https://doi.org/10.1038/s41569-021-00549-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-021-00549-w
This article is cited by
-
A call for increased inclusivity and global representation in pharmacogenetic testing
npj Genomic Medicine (2024)
-
The Impact of CYP2C19 Genotype on the Platelet Reactivity Index (PRI) among Chronic Coronary Syndromes (CCS) Patients Undergoing Percutaneous Coronary Intervention (PCI): Affectability of Rapid Genetic Testing
Cardiovascular Drugs and Therapy (2024)
-
Human Serum Albumin-enriched Clopidogrel Bisulfate Nanoparticle Alleviates Cerebral Ischemia–Reperfusion Injury in Rats
Pharmaceutical Research (2023)
-
Pharmacogenomics implementation in cardiovascular disease in a highly diverse population: initial findings and lessons learned from a pilot study in United Arab Emirates
Human Genomics (2022)
-
Transcriptomic data exploration of consensus genes and molecular mechanisms between chronic obstructive pulmonary disease and lung adenocarcinoma
Scientific Reports (2022)