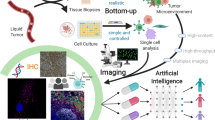
Abstract
Tissue imaging has become much more colourful in the past decade. Advances in both experimental and analytical methods now make it possible to image protein markers in tissue samples in high multiplex. The ability to routinely image 40–50 markers simultaneously, at single-cell or subcellular resolution, has opened up new vistas in the study of tumour biology. Cellular phenotypes, interaction, communication and spatial organization have become amenable to molecular-level analysis, and application to patient cohorts has identified clinically relevant cellular and tissue features in several cancer types. Here, we review the use of multiplex protein imaging methods to study tumour biology, discuss ongoing attempts to combine these approaches with other forms of spatial omics, and highlight challenges in the field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). The second iteration of this seminal review of the hallmarks of cancer, synthesizing evidence for immune evasion, inflammation and the TME as important features of the disease.
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Tabassum, D. P. & Polyak, K. Tumorigenesis: it takes a village. Nat. Rev. Cancer 15, 473–483 (2015).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280.e26 (2021).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Heppner, G. H. Tumor heterogeneity. Cancer Res. 44, 2259–2265 (1984).
Bi, K. et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 39, 649–661.e5 (2021).
Bhaduri, A. et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 26, 48–63.e6 (2020).
Wang, L. et al. A single-cell atlas of glioblastoma evolution under therapy reveals cell-intrinsic and cell-extrinsic therapeutic targets. Nat. Cancer 3, 1534–1552 (2022).
Vegliante, R., Pastushenko, I. & Blanpain, C. Deciphering functional tumor states at single-cell resolution. EMBO J. 41, e109221 (2022).
Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. & Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349–1360 (2018).
Izar, B. et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 26, 1271–1279 (2020).
Massalha, H. et al. A single cell atlas of the human liver tumor microenvironment. Mol. Syst. Biol. 16, e9682 (2020).
Zhou, Y. et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 11, 6322 (2020).
Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809.e23 (2021).
Chen, Y. et al. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat. Commun. 13, 4851 (2022).
Nieto, P. et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 31, 1913–1926 (2021).
Bolis, M. et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat. Commun. 12, 7033 (2021).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Kumar, V. et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 12, 670–691 (2022).
Cords, L. et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 14, 4294 (2023).
Tietscher, S. et al. A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer. Nat. Commun. 14, 98 (2023).
Chijimatsu, R. et al. Establishment of a reference single-cell RNA sequencing dataset for human pancreatic adenocarcinoma. iScience 25, 104659 (2022).
Chu, Y. et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat. Med. 29, 1550–1562 (2023).
Rozenblatt-Rosen, O. et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Lewis, S. M. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021).
Elhanani, O., Ben-Uri, R. & Keren, L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 41, 404–420 (2023).
de Vries, N. L., Mahfouz, A., Koning, F. & de Miranda, N. Unraveling the complexity of the cancer microenvironment with multidimensional genomic and cytometric technologies. Front. Oncol. 10, 1254 (2020).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018). A MIBI study of triple-negative breast cancer that defines immune cell patterns, immunoregulatory protein expression and spatial features of tumour tissue.
Danenberg, E. et al. Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat. Genet. 54, 660–669 (2022). An IMC study of the TME in breast cancer that identifies spatial patterns associated with specific driver mutations and with patient survival.
Sorin, M. et al. Single-cell spatial landscapes of the lung tumour immune microenvironment. Nature 614, 548–554 (2023).
Karimi, E. et al. Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature 614, 555–563 (2023).
Ferguson, A. L. et al. High-dimensional and spatial analysis reveals immune landscape-dependent progression in cutaneous squamous cell carcinoma. Clin. Cancer Res. 28, 4677–4688 (2022).
Xiao, X. et al. Multiplexed imaging mass cytometry reveals distinct tumor-immune microenvironments linked to immunotherapy responses in melanoma. Commun. Med. 2, 131 (2022).
Hoch, T. et al. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 7, eabk1692 (2022). A RNA-IMC study of metastatic melanoma that identifies local patches of chemokine-secreting cells together with the phenotypes of the cells in the surrounding milieus.
Lin, J. R. et al. Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer. Cell 186, 363–381.e19 (2023). A CycIF study of colorectal cancer that identifies molecular features underlying large-scale histological patterns and highlights the limitations of imaging small fields of view such as those in tumour microarrays.
Matusiak, M. et al. A spatial map of human macrophage niches reveals context-dependent macrophage functions in colon and breast cancer. Res. Sq. https://doi.org/10.21203/rs.3.rs-2393443/v1 (2023).
Hartmann, F. J. et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 39, 186–197 (2021). A MIBI and mass cytometry study that examines metabolic programmes of different cell types in colorectal cancer and maps their spatial organization.
Remark, R. et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 1, aaf6925 (2016).
Tsujikawa, T. et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 19, 203–217 (2017).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Banik, G. et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Methods Enzymol. 635, 1–20 (2020).
Toki, M. I. et al. High-plex predictive marker discovery for melanoma immunotherapy-treated patients using digital spatial profiling. Clin. Cancer Res. 25, 5503–5512 (2019).
Schulz, D. et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 6, 25–36.e5 (2018).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Gaglia, G. et al. Temporal and spatial topography of cell proliferation in cancer. Nat. Cell Biol. 24, 316–326 (2022).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020). An IMC study of breast cancer showing that multiplex imaging-defined single-cell pathology groups correlate with patient survival beyond the information provided by clinical subtypes.
Ali, H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 1, 163–175 (2020).
Fischer, J. R. et al. Multiplex imaging of breast cancer lymph node metastases identifies prognostic single-cell populations independent of clinical classifiers. Cell Rep. Med. 4, 100977 (2023). An IMC study showing cell phenotypic divergence between paired primary breast tumours and lymph node metastases, and identifying prognostic phenotypes in metastatic tumours.
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Wang, X. Q. et al. Spatial predictors of immunotherapy response in triple-negative breast cancer. Nature 621, 868–876 (2023). An IMC study conducted on a cohort of patients with breast cancer within a clinical trial and identifying cell phenotypic and spatial predictors of immunotherapy response.
Lin, J. R. et al. High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers. Nat. Cancer 4, 1036–1052 (2023).
Radtke, A. J. et al. A multi-scale, multi-omic atlas of human normal and follicular lymphoma lymph nodes. Preprint at bioRxiv https://doi.org/10.1101/2022.06.03.494716 (2022).
Milosevic, V. Different approaches to imaging mass cytometry data analysis. Bioinform Adv. 3, vbad046 (2023).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022).
Zhang, M. et al. Spatial molecular profiling: platforms, applications and analysis tools.Brief. Bioinform. 22, bbaa145 (2021).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020). A CODEX study on colorectal cancer illustrating the definition of cellular neighbourhoods and showing that spatial organization differs in samples with and without TLSs.
Chen, Z., Soifer, I., Hilton, H., Keren, L. & Jojic, V. Modeling multiplexed images with spatial-LDA reveals novel tissue microenvironments. J. Comput. Biol. 27, 1204–1218 (2020).
Phillips, D. et al. Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nat. Commun. 12, 6726 (2021). A CODEX study on cutaneous T cell lymphoma showing that spatial organization but not cell phenotypes is associated with patient response to immunotherapy.
Bhate, S. S., Barlow, G. L., Schürch, C. M. & Nolan, G. P. Tissue schematics map the specialization of immune tissue motifs and their appropriation by tumors. Cell Syst. 13, 109–130.e6 (2022).
Damond, N. et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 29, 755–768.e55 (2019).
Risom, T. et al. Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell 185, 299–310.e18 (2022). A MIBI study of early-stage breast cancer that used histological staining to define areas of interest for multiplex imaging and identifies features prognostic for disease progression.
Hickey, J. W. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods 19, 284–295 (2022). A detailed practical guide to many aspects of multiplex protein imaging.
Moffitt, J. R., Lundberg, E. & Heyn, H. The emerging landscape of spatial profiling technologies. Nat. Rev. Genet. 23, 741–759 (2022).
Windhager, J. et al. An end-to-end workflow for multiplexed image processing and analysis. Nat. Protoc. 18, 3565–3613 (2023).
Liu, C. C. et al. Multiplexed ion beam imaging: insights into pathobiology. Annu. Rev. Pathol. 17, 403–423 (2022).
Kuswanto, W., Nolan, G. & Lu, G. Highly multiplexed spatial profiling with CODEX: bioinformatic analysis and application in human disease. Semin. Immunopathol. 45, 145–157 (2023).
Baharlou, H., Canete, N. P., Cunningham, A. L., Harman, A. N. & Patrick, E. Mass cytometry imaging for the study of human diseases-applications and data analysis strategies. Front. Immunol. 10, 2657 (2019).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014). The first methodological report of MIBI applied to human breast tumours.
Keren, L. et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 5, eaax5851 (2019).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). The first methodological report of IMC applied to human breast tumours.
Jiang, S. et al. Combined protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments. Immunity 55, 1118–1134.e8 (2022).
Kuett, L. et al. Three-dimensional imaging mass cytometry for highly multiplexed molecular and cellular mapping of tissues and the tumor microenvironment. Nat. Cancer 3, 122–133 (2022).
Hosogane, T., Casanova, R. & Bodenmiller, B. DNA-barcoded signal amplification for imaging mass cytometry enables sensitive and highly multiplexed tissue imaging. Nat. Methods 20, 1304–1309 (2023).
Rovira-Clavé, X. et al. Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging. Nat. Commun. 12, 4628 (2021).
Goossens, P. et al. Integrating multiplex immunofluorescent and mass spectrometry imaging to map myeloid heterogeneity in its metabolic and cellular context. Cell Metab. 34, 1214–1225.e6 (2022).
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012).
Radtke, A. J. et al. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020). A methodological paper describing the rapid, flexible and highly-multiplex IBEX method for cyclic immunofluorescence imaging.
Schubert, W. et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 24, 1270–1278 (2006).
Lin, J. R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018). An early methodological report of highly multiplex CycIF applied to tissue.
Wählby, C., Erlandsson, F., Bengtsson, E. & Zetterberg, A. Sequential immunofluorescence staining and image analysis for detection of large numbers of antigens in individual cell nuclei. Cytometry 47, 32–41 (2002). The first multiplex imaging paper, showing that immunofluorescence signals can be sequentially removed without destroying antigenicity in fixed and paraffin-embedded tissue.
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042 (2018).
Gaglia, G. et al. Lymphocyte networks are dynamic cellular communities in the immunoregulatory landscape of lung adenocarcinoma. Cancer Cell 41, 871–886.e10 (2023).
Nirmal, A. J. et al. The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Cancer Discov. 12, 1518–1541 (2022).
Li, K. et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti-PD-1 therapy. Cancer Cell 40, 1374–1391.e7 (2022).
Chen, M. et al. Spatiotemporal analysis of B cell- and antibody secreting cell-subsets in human melanoma reveals metastasis-, tumor stage-, and age-associated dynamics. Front. Cell Dev. Biol. 9, 677944 (2021).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Black, S. et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021). A methodological paper describing the CODEX workflow.
Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01676-0 (2023).
Ben-Chetrit, N. et al. Integration of whole transcriptome spatial profiling with protein markers. Nat. Biotechnol. 41, 788–793 (2023).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Kishi, J. Y. et al. Light-Seq: light-directed in situ barcoding of biomolecules in fixed cells and tissues for spatially indexed sequencing. Nat. Methods 19, 1393–1402 (2022).
Hu, K. H. et al. ZipSeq: barcoding for real-time mapping of single cell transcriptomes. Nat. Methods 17, 833–843 (2020).
Sha, L. et al. Integrated spatial transcriptomic and proteomic analysis of fresh frozen tissue based on stereo-seq. Preprint at: bioRxiv https://doi.org/10.1101/2023.04.28.538364 (2023).
McNamara, K. L. et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat. Cancer 2, 400–413 (2021).
Brady, L. et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 12, 1426 (2021).
Martinez-Morilla, S. et al. Digital spatial profiling of melanoma shows CD95 expression in immune cells is associated with resistance to immunotherapy. Oncoimmunology 12, 2260618 (2023).
Gavrielatou, N. et al. Digital spatial profiling links beta-2-microglobulin expression with immune checkpoint blockade outcomes in head and neck squamous cell carcinoma. Cancer Res. Commun. 3, 558–563 (2023).
Schoenfeld, D. A. et al. Immune dysfunction revealed by digital spatial profiling of immuno-oncology markers in progressive stages of renal cell carcinoma and in brain metastases. J. Immunother. Cancer 11, e007240 (2023).
Carter, J. M. et al. Distinct spatial immune microlandscapes are independently associated with outcomes in triple-negative breast cancer. Nat. Commun. 14, 2215 (2023).
Bonnett, S. A. et al. Ultra high-plex spatial proteogenomic investigation of giant cell glioblastoma multiforme immune infiltrates reveals distinct protein and RNA expression profiles. Cancer Res. Commun. 3, 763–779 (2023).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Govek, K. W. et al. Single-cell transcriptomic analysis of mIHC images via antigen mapping. Sci. Adv. 7, eabc5464 (2021).
Zhu, B. et al. Robust single-cell matching and multimodal analysis using shared and distinct features. Nat. Methods 20, 304–315 (2023).
Chen, S. et al. Integration of spatial and single-cell data across modalities with weakly linked features. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01935-0 (2023).
Strand, S. H. et al. Molecular classification and biomarkers of clinical outcome in breast ductal carcinoma in situ: analysis of TBCRC 038 and RAHBT cohorts. Cancer Cell 40, 1521–1536.e7 (2022).
Chan, J. M. et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39, 1479–1496.e18 (2021).
Vázquez-García, I. et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 612, 778–786 (2022).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497–514.e22 (2020).
Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e13 (2022).
Müller, W. H., De Pauw, E., Far, J., Malherbe, C. & Eppe, G. Imaging lipids in biological samples with surface-assisted laser desorption/ionization mass spectrometry: a concise review of the last decade. Prog. Lipid Res. 83, 101114 (2021).
Balluff, B., Hanselmann, M. & Heeren, R. M. Mass spectrometry imaging for the investigation of intratumor heterogeneity. Adv. Cancer Res. 134, 201–230 (2017).
Ma, X. & Fernández, F. M. Advances in mass spectrometry imaging for spatial cancer metabolomics. Mass Spectrom. Rev. 2022, e21804 (2022).
Spraggins, J. M. et al. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem. 91, 14552–14560 (2019).
Niehaus, M., Soltwisch, J., Belov, M. E. & Dreisewerd, K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 16, 925–931 (2019).
Zavalin, A., Yang, J., Hayden, K., Vestal, M. & Caprioli, R. M. Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal. Bioanal. Chem. 407, 2337–2342 (2015).
Neumann, E. K., Comi, T. J., Rubakhin, S. S. & Sweedler, J. V. Lipid heterogeneity between astrocytes and neurons revealed by single-cell MALDI-MS combined with immunocytochemical classification. Angew. Chem. Int. Ed. Engl. 58, 5910–5914 (2019).
Cuypers, E. et al. ‘On the Spot’ digital pathology of breast cancer based on single-cell mass spectrometry imaging. Anal. Chem. 94, 6180–6190 (2022).
Bien, T. et al. MALDI-2 mass spectrometry and immunohistochemistry imaging of Gb3Cer, Gb4Cer, and further glycosphingolipids in human colorectal cancer tissue. Anal. Chem. 92, 7096–7105 (2020).
O’Neill, K. C., Liapis, E., Harris, B. T., Perlin, D. S. & Carter, C. L. Mass spectrometry imaging discriminates glioblastoma tumor cell subpopulations and different microvascular formations based on their lipid profiles. Sci. Rep. 12, 17069 (2022).
Andersen, M. K. et al. Spatial differentiation of metabolism in prostate cancer tissue by MALDI-TOF MSI. Cancer Metab. 9, 9 (2021).
Ščupáková, K. et al. Clinical importance of high-mannose, fucosylated, and complex N-glycans in breast cancer metastasis. JCI Insight 6, e146945 (2021).
Prentice, B. M. et al. Imaging mass spectrometry enables molecular profiling of mouse and human pancreatic tissue. Diabetologia 62, 1036–1047 (2019).
Prade, V. M. et al. De novo discovery of metabolic heterogeneity with immunophenotype-guided imaging mass spectrometry. Mol. Metab. 36, 100953 (2020).
Wang, G. et al. Analyzing cell-type-specific dynamics of metabolism in kidney repair. Nat. Metab. 4, 1109–1118 (2022).
Baker, E. A. G., Schapiro, D., Dumitrascu, B., Vickovic, S. & Regev, A. In silico tissue generation and power analysis for spatial omics. Nat. Methods 20, 424–431 (2023).
Bost, P., Schulz, D., Engler, S., Wasserfall, C. & Bodenmiller, B. Optimizing multiplexed imaging experimental design through tissue spatial segregation estimation. Nat. Methods 20, 418–423 (2023).
Edfors, F. et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 12, 883 (2016).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). A paper discussing the imperfect relationship between transcript and protein levels and arguing that protein-level measurements are needed to understand biological systems in many contexts.
Du, Z. et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat. Protoc. 14, 2900–2930 (2019). A methodological paper that describes a pipeline for antibody validation.
Uhlen, M. et al. A proposal for validation of antibodies. Nat. Methods 13, 823–827 (2016).
Quardokus, E. M. et al. Organ mapping antibody panels: a community resource for standardized multiplexed tissue imaging. Nat. Methods 20, 1174–1178 (2023).
Liu, C. C. et al. Robust phenotyping of highly multiplexed tissue imaging data using pixel-level clustering. Nat. Commun. 14, 4618 (2023).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Spitzer, H., Berry, S., Donoghoe, M., Pelkmans, L. & Theis, F. J. Learning consistent subcellular landmarks to quantify changes in multiplexed protein maps. Nat. Methods https://doi.org/10.1038/s41592-023-01894-z (2023).
Bai, Y. et al. Expanded vacuum-stable gels for multiplexed high-resolution spatial histopathology. Nat. Commun. 14, 4013 (2023).
Strotton, M. et al. Multielement Z-tag imaging by X-ray fluorescence microscopy for next-generation multiplex imaging. Nat. Methods 20, 1310–1322 (2023).
Yagnik, G., Liu, Z., Rothschild, K. J. & Lim, M. J. Highly multiplexed immunohistochemical MALDI-MS imaging of biomarkers in tissues. J. Am. Soc. Mass Spectrom. 32, 977–988 (2021).
Claes, B. S. R. et al. MALDI-IHC-guided in-depth spatial proteomics: targeted and untargeted MSI combined. Anal. Chem. 95, 2329–2338 (2023).
Wei, L. et al. Super-multiplex vibrational imaging. Nature 544, 465–470 (2017).
Shi, L. et al. Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nat. Biotechnol. 40, 364–373 (2022).
Wu, E. et al. 7-UP: generating in silico CODEX from a small set of immunofluorescence markers. PNAS Nexus 2, pgad171 (2023).
Ben-Uri, R. et al. Escalating high-dimensional imaging using combinatorial channel multiplexing and deep learning. Preprint at: bioRxiv https://doi.org/10.1101/2023.09.09.556962 (2023).
Zhang, W. et al. Identification of cell types in multiplexed in situ images by combining protein expression and spatial information using CELESTA. Nat. Methods 19, 759–769 (2022).
Brbić, M. et al. Annotation of spatially resolved single-cell data with STELLAR. Nat. Methods 19, 1411–1418 (2022).
Amitay, Y. et al. CellSighter: a neural network to classify cells in highly multiplexed images. Nat. Commun. 14, 4302 (2023).
Mund, A. et al. Deep visual proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40, 1231–1240 (2022).
Cui, Y. et al. Expansion microscopy using a single anchor molecule for high-yield multiplexed imaging of proteins and RNAs. PLoS One 18, e0291506 (2023).
Ghazanfar, S., Guibentif, C. & Marioni, J. C. Stabilized mosaic single-cell data integration using unshared features. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01766-z (2023).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01767-y (2023).
Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, eabc4552 (2021).
Cullin, N., Azevedo Antunes, C., Straussman, R., Stein-Thoeringer, C. K. & Elinav, E. Microbiome and cancer. Cancer Cell 39, 1317–1341 (2021).
Galeano Niño, J. L. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022).
Ozeki, M. et al. Susceptibility of actin to modification by 4-hydroxy-2-nonenal. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 827, 119–126 (2005).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022).
Montanari, N. R. et al. Multi-parametric analysis of human livers reveals variation in intrahepatic inflammation across phases of chronic hepatitis B infection. J. Hepatol. 77, 332–343 (2022).
Hoyt, C. C. Multiplex immunofluorescence and multispectral imaging: forming the basis of a clinical test platform for immuno-oncology. Front. Mol. Biosci. 8, 674747 (2021).
Hurov, K. et al. BT7480, a novel fully synthetic Bicycle tumor-targeted immune cell agonist™ (Bicycle TICA™) induces tumor localized CD137 agonism. J. Immunother. Cancer 9, e002883 (2021).
Rivest, F. et al. Fully automated sequential immunofluorescence (seqIF) for hyperplex spatial proteomics. Sci. Rep. 13, 16994 (2023).
Jarosch, S. et al. Multiplexed imaging and automated signal quantification in formalin-fixed paraffin-embedded tissues by ChipCytometry. Cell Rep. Methods 1, 100104 (2021).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Peng, T. et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8, 14836 (2017).
Lu, P. et al. IMC-Denoise: a content aware denoising pipeline to enhance imaging mass cytometry. Nat. Commun. 14, 1601 (2023).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Schapiro, D. et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat. Methods 19, 311–315 (2022).
Marconato, L. et al. SpatialData: an open and universal data framework for spatial omics. Preprint at bioRxiv https://doi.org/10.1101/2023.05.05.539647 (2023).
Acknowledgements
We thank Nils Eling and Daniel Schulz for critical reading and feedback on the manuscript. We thank all Bodenmiller laboratory members for helpful discussions. B.B. was funded by two SNSF grants (310030_205007, 316030_213512), an NIH grant (UC4 DK108132), the CRUK IMAXT Grand Challenge, and the European Research Council (ERC) under the European Union’s Horizon 2020 Program under the ERC grant agreement no. 866074 (“Precision Motifs”).
Author information
Authors and Affiliations
Contributions
N.d.S. and S.Z. researched data for the article. All authors contributed substantially to discussion of the content. N.d.S. wrote and revised the article. S.Z. prepared and revised the figures and tables with input from the other authors. N.d.S. and B.B. reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
B.B. has founded and is a shareholder and member of the board of Navignostics, a precision oncology spin-off from the University of Zurich. N.d.S. and S.Z. declare no competing interests.
Peer review
Peer review information
Nature Reviews Cancer thanks Sizun Jiang who co-reviewed with Hendrik Michel, Christian Schürch and Sean Bendall for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Human protein atlas: https://www.proteinatlas.org/
Napari: https://zenodo.org/record/7276432
Online documentation of IMC data analysis: https://bodenmillergroup.github.io/IMCDataAnalysis/
Supplementary information
Glossary
- Cell segmentation
-
An image processing step that delineates the boundaries of individual cells.
- Chromogen
-
A compound that can be converted into a coloured product that can be detected by light microscopy.
- Co-detection by indexing
-
(CODEX). A highly multiplex protein imaging technique that uses iterative hybridization and stripping of fluorophore-tagged DNA oligonucleotide probes to image samples stained with DNA barcode-tagged antibodies.
- Cyclic immunofluorescence
-
(CycIF). A highly multiplex imaging technique using iterative staining with fluorophore-tagged antibodies coupled with chemical inactivation of fluorophores between staining cycles to build a multiplex image of a labelled sample.
- Digital spatial profiling
-
(DSP). A highly multiplex profiling technique for mRNA or protein that uses patterned light to release UV-photocleavable oligonucleotide tags attached to antibodies or to RNA probes in a defined spatial region, followed by sequencing or single molecule counting as a readout.
- Dimensionality reduction
-
A data-processing approach whereby high-dimensional data are projected into a low number of dimensions represented by a smaller subset of variables with essentially the same information content as the full measured set.
- Expansion microscopy
-
An approach that uses polymer-based physical expansion of a sample to improve the resolution of fluorescence microscopy beyond the diffraction limit of light.
- Haptens
-
Small molecules that are not intrinsically antigenic but become so in combination with a macromolecule such as a protein.
- Image registration
-
Data processing steps that bring two or more different images into a single coordinate system such that the images can be aligned.
- Imaging mass cytometry
-
(IMC). A highly multiplex protein or RNA imaging technique that couples mass cytometry by time of flight with high-resolution laser ablation to image samples labelled with metal isotope reporter-tagged antibodies.
- Imputation
-
An approach for handling missing data, typically by replacement with substitute values.
- Multiplexed ion beam imaging
-
(MIBI). A highly multiplex protein imaging technique that uses secondary ion mass spectrometry to image samples labelled with metal isotope reporter-tagged antibodies.
- Raman microscopy
-
Spatially resolved chemical analysis of a sample based on the detection of vibrational modes by scattered light.
- Signal amplification by exchange reaction
-
(SABER). A signal amplification approach based on hybridization of imager DNA strands to concatemerized DNA barcodes assembled on antibodies used to label a sample; compatible with fluorescence and mass cytometric multiplex imaging.
- Spillover correction
-
Data processing steps that compensate for fluorescent or metal signals from one channel that are detected artefactually in a different channel.
- Synchrotron
-
A machine that accelerates charged particles (electrons) to almost the speed of light and thereby generates very intense light, mostly in the X-ray region.
- Tertiary lymphoid structures
-
(TLS). Structured multicellular aggregates of immune cells found outside of lymph nodes, in peripheral tissue, and that reflect inflammatory signalling in the tissue.
- Tyramide-based amplification
-
A signal amplification approach in which horseradish peroxidase (typically coupled to an antibody) catalyses the conversion of labelled tyramide to a reactive molecule that covalently labels nearby proteins at high density.
- Voxel gating
-
Data processing steps to identify a cell of interest based on selecting voxels that are positive for expected markers and negative for incorrect or irrelevant markers.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Souza, N., Zhao, S. & Bodenmiller, B. Multiplex protein imaging in tumour biology. Nat Rev Cancer 24, 171–191 (2024). https://doi.org/10.1038/s41568-023-00657-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-023-00657-4