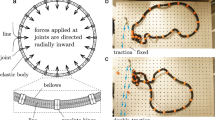
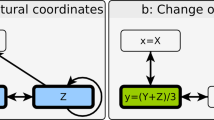
Abstract
Understanding conformational change is crucial for programming and controlling the function of many mechanobiological and mechanical systems such as robots, enzymes and tunable metamaterials. These systems are often modelled as constituent nodes (for example, joints or amino acids) whose motion is restricted by edges (for example, limbs or bonds) to yield functionally useful coordinated motions (for example, walking or allosteric regulation). However, the design of desired functions is made difficult by the complex dependence of these coordinated motions on the connectivity of edges. Here, we develop simple mathematical principles to design mechanical systems that achieve any desired infinitesimal or finite coordinated motion. We specifically study mechanical networks of two- and three-dimensional frames composed of nodes connected by freely rotating rods and springs. We first develop simple principles that govern all networks with an arbitrarily specified motion as the sole zero-energy mode. We then extend these principles to characterize networks that yield multiple specified zero modes, generate pre-stress stability and display branched motions. By coupling individual modules, we design networks with negative Poisson’s ratio and allosteric response. Finally, we extend our framework to networks with arbitrarily specifiable initial and final positions to design energy minima at desired geometric configurations, and create networks demonstrating tristability and cooperativity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
There are no data with mandated deposition used in the manuscript or Supplementary Information. All analyses and figures were created in MATLAB, and can be publicly accessed on GitHub at https://github.com/jk6294/Mechanical-Networks.git with a test script that will exactly replicate most of the key figures and results in this manuscript.
References
Papadopoulos, L., Porter, M. A., Daniels, K. E. & Bassett, D. S. Network analysis of particles and grains. J. Complex Netw. 6, 485–565 (2018).
Picu, R. C. Mechanics of random fiber networks—a review. Soft Matter 7, 6768–6785 (2011).
Vermeulen, M. F. J., Bose, A., Storm, C. & Ellenbroek, W. G. Geometry and the onset of rigidity in a disordered network. Phys. Rev. E 96, 053003 (2017).
Bassett, D. S., Owens, E. T., Daniels, K. E. & Porter, M. A. Influence of network topology on sound propagation in granular materials. Phys. Rev. E 86, 041306 (2012).
Shi, F., Wang, S., Forest, M. G. & Mucha, P. J. Network-based assessments of percolation-induced current distributions in sheared rod macromolecular dispersions. Multiscale Model. Simul. 12, 249–264 (2014).
Detweiler, C., Vona, M., Yoon, Y., Yun, Seung-Kook & Rus, D. Self-assembling mobile linkages. IEEE Robotics Autom. Mag. 14, 45–55 (2007).
Patek, S. N., Nowroozi, B. N., Baio, J. E., Caldwell, R. L. & Summers, A. P. Linkage mechanics and power amplification of the mantis shrimp’s strike. J. Exp. Biol. 210, 3677–3688 (2007).
Crapo, H. Structural rigidity. Struct. Topol. 73, 26–45 (1979).
Maxwell, J. C. On the calculation of the equilibrium and stiffness of frames. Phil. Maga. Ser. 4 27, 294–299 (1864).
Grimm, H. & Dorner, B. On the mechanism of the α-β phase transformation of quartz. J. Phys. Chem. Solids 36, 407–413 (1975).
Bertoldi, K., Vitelli, V., Christensen, J. & van Hecke, M. Flexible mechanical metamaterials. Nat. Rev. Mater. 2, 17066 (2017).
Guo, J. & Zhou, H. X. Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 (2016).
Kempe, A. B. On a general method of describing plane curves of the nth degree by linkwork. Proc. Lond. Math. Soc. s1–7, 213–216 (1875).
Goodrich, C. P., Liu, A. J. & Nagel, S. R. The principle of independent bond-level response: tuning by pruning to exploit disorder for global behavior. Phys. Rev. Lett. 114, 225501 (2015).
Bolker, E. & Roth, B. When is a bipartite graph a rigid framework? Pac. J. Math. 90, 27–44 (1980).
Guest, S. The stiffness of prestressed frameworks: a unifying approach. Int. J. Solids Struct. 43, 842–854 (2006).
Asimow, L. & Roth, B. The rigidity of graphs. Trans. Am. Math. Soc. 245, 279–289 (1978).
Changeux, J.-P. & Edelstein, S. J. Allosteric receptors after 30 years. Neuron 21, 959–980 (1998).
Allewell, N. M. Escherichia coli aspartate transcarbamoylase: structure, energetics, and catalytic and regulatory mechanisms. Annu. Rev. Biophys. Biophys. Chem. 18, 71–92 (1989).
Macol, C. P., Tsuruta, H., Stec, B. & Kantrowitz, E. R. Direct structural evidence for a concerted allosteric transition in Escherichia coli aspartate transcarbamoylase. Nat. Struct. Biol. 8, 423–426 (2001).
Cockrell, G. M. et al. New paradigm for allosteric regulation of Escherichia coli aspartate transcarbamoylase. Biochemistry 52, 8036–8047 (2013).
Rocks, J. W. et al. Designing allostery-inspired response in mechanical networks. Proc. Natl Acad. Sci. USA 114, 2520–2525 (2017).
Whiteley, W. Infinitesimal motions of a bipartite framework. Pac. J. Math. 110, 233–255 (1984).
Silverberg, J. L. et al. Using origami design principles to fold reprogrammable mechanical metamaterials. Science 345, 647–650 (2014).
Paulose, J., Chen, B. G.-g & Vitelli, V. Topological modes bound to dislocations in mechanical metamaterials. Nat. Phys. 11, 153–156 (2015).
Körner, C. & Liebold-Ribeiro, Y. A systematic approach to identify cellular auxetic materials. Smart Mater. Struct. 24, 025013 (2015).
Lukin, J. A. & Ho, C. The structure function relationship of hemoglobin in solution at atomic resolution. Chem. Rev. 104, 1219–1230 (2004).
Lee, J.-H., Singer, J. P. & Thomas, E. L. Micro-/nanostructured mechanical metamaterials. Adv. Mater. 24, 4782–4810 (2012).
Dagdelen, J., Montoya, J., de Jong, M. & Persson, K. Computational prediction of new auxetic materials. Nat. Commun. 8, 323 (2017).
Yu, Z. & Qian, W.-H. Dynamic force distribution in multifingered grasping by decomposition and positive combination. IEEE Trans. Robotics 21, 718–726 (2005).
Acknowledgements
We gratefully acknowledge conversations with B. Chen, A. E. Sizemore, E. J. Cornblath, E. Teich and M. X. Lim. J.Z.K. acknowledges support from the NIH T32-EB020087, PD: F. W. Wehrli, and the National Science Foundation Graduate Research Fellowship no. DGE-1321851. S.H.S. acknowledges support from the United States NSF grant nos. DMS-1513179 and CCF-1522054. D.S.B. acknowledges support from the John D. and Catherine T. MacArthur Foundation, the ISI Foundation, the Alfred P. Sloan Foundation, an NSF CAREER award PHY-1554488, and from the NSF through the University of Pennsylvania Materials Research Science and Engineering Center (MRSEC) DMR-1720530.
Author information
Authors and Affiliations
Contributions
J.Z.K. and D.S.B. wrote the manuscript, with feedback from Z.L. and S.H.S. J.Z.K. conceived the idea, formalized the math, and performed the simulations and analyses with feedback from Z.L., S.H.S. and D.S.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J.Z., Lu, Z., Strogatz, S.H. et al. Conformational control of mechanical networks. Nat. Phys. 15, 714–720 (2019). https://doi.org/10.1038/s41567-019-0475-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-019-0475-y
This article is cited by
-
Impact of physicality on network structure
Nature Physics (2024)
-
Geometry for mechanics
Nature Physics (2019)