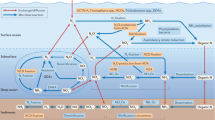
Abstract
One-quarter of photosynthesis-derived carbon on Earth rapidly cycles through a set of short-lived seawater metabolites that are generated from the activities of marine phytoplankton, bacteria, grazers and viruses. Here we discuss the sources of microbial metabolites in the surface ocean, their roles in ecology and biogeochemistry, and approaches that can be used to analyse them from chemistry, biology, modelling and data science. Although microbial-derived metabolites account for only a minor fraction of the total reservoir of marine dissolved organic carbon, their flux and fate underpins the central role of the ocean in sustaining life on Earth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Azam, F. et al. The ecological role of water-column microbes. Mar. Ecol. Prog. Ser. 10, 257–263 (1983).
Hansell, D. A. Recalcitrant dissolved organic carbon fractions. Ann. Rev. Mar. Sci. 5, 421–445 (2013).
Weber, L. et al. Extracellular reef metabolites across the protected Jardines de la Reina, Cuba reef system. Front. Mar. Sci. https://doi.org/10.3389/fmars.2020.582161 (2020).
Widner, B., Kido Soule, M. C., Ferrer-González, F. X., Moran, M. A. & Kujawinski, E. B. Quantification of amine-and alcohol-containing metabolites in saline samples using pre-extraction benzoyl chloride derivatization and ultrahigh performance liquid chromatography tandem mass spectrometry (UHPLC MS/MS). Anal. Chem. 93, 4809–4817 (2021).
Fogg, G. E. The ecological significance of extracellular products of phytoplankton photosynthesis. Bot. Mar. 26, 3–14 (1983).
Morán, X. A. G., Ducklow, H. W. & Erickson, M. Carbon fluxes through estuarine bacteria reflect coupling with phytoplankton. Mar. Ecol. Prog. Ser. 489, 75–85 (2013).
Mühlenbruch, M., Grossart, H. P., Eigemann, F. & Voss, M. Phytoplankton‐derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ. Microbiol. 20, 2671–2685 (2018).
Flynn, K. J., Clark, D. R. & Xue, Y. Modeling the release of dissolved organic matter by phytoplankton 1. J. Phycol. 44, 1171–1187 (2008).
Bjørrisen, P. K. Phytoplankton exudation of organic matter: why do healthy cells do it? Limnol. Oceanogr. 33, 151–154 (1988).
Myklestad, S. M. Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci. Total Environ. 165, 155–164 (1995).
Falkowski, P. & Raven, J. Aquatic Photosynthesis 2nd edn (Princeton Univ. Press, 2007).
Lau, W. W. & Armbrust, E. V. Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ. Microbiol. 8, 1688–1702 (2006).
Wood, A. M. & Van Valen, L. M. Paradox lost? On the release of energy-rich compounds by photoplankton. Mar. Microb. Food Webs 4, 103–116 (1990).
Hellebust, J. Excretion of organic compounds by cultured and natural populations of marine phytoplankton. Estuaries 10, 192–206 (1967).
Thornton, D. C. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46 (2014).
Wangersky, P. in Micobial Ecology Vol. IV (ed. Kinne, O) 115–220 (Wiley, 1978).
Braakman, R., Follows, M. J. & Chisholm, S. W. Metabolic evolution and the self-organization of ecosystems. Proc. Natl Acad. Sci. USA 114, E3091–E3100 (2017).
Durham, B. P. et al. Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat. Microbiol. 4, 1706–1715 (2019).
Mague, T., Friberg, E., Hughes, D. & Morris, I. Extracellular release of carbon by marine phytoplankton; a physiological approach. Limnol. Oceanogr. 24, 262–279 (1980).
Vardi, A. et al. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 4, e60 (2006).
Gillard, J. et al. Metabolomics enables the structure elucidation of a diatom sex pheromone. Angew. Chem. Int. Ed. 52, 854–857 (2013).
Brunson, J. K. et al. Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 361, 1356–1358 (2018).
Legrand, C., Rengefors, K., Fistarol, G. O. & Graneli, E. Allelopathy in phytoplankton-biochemical, ecological and evolutionary aspects. Phycologia 42, 406–419 (2003).
Granum, E., Kirkvold, S. & Myklestad, S. M. Cellular and extracellular production of carbohydrates and amino acids by the marine diatom Skeletonema costatum: diel variations and effects of N depletion. Mar. Ecol. Prog. Ser. 242, 83–94 (2002).
Mague, T., Friberg, E., Hughes, D. & Morris, I. Extracellular release of carbon by marine phytoplankton; a physiological approach 1. Limnol. Oceanogr. 25, 262–279 (1980).
Puskaric, S. & Mortain-Bertrand, A. Physiology of diatom Skeletonema costatum (Grev.) Cleve photosynthetic extracellular release: evidence for a novel coupling between marine bacteria and phytoplankton. J. Plankton Res. 25, 1227–1235 (2003).
Aylward, F. O. et al. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc. Natl Acad. Sci. USA 112, 5443–5448 (2015).
Frischkorn, K. R., Haley, S. T. & Dyhrman, S. T. Coordinated gene expression between Trichodesmium and its microbiome over day–night cycles in the North Pacific Subtropical Gyre. ISME J. 12, 997–1007 (2018).
Bidle, K. D. The molecular ecophysiology of programmed cell death in marine phytoplankton. Ann. Rev. Mar. Sci. 7, 341–375 (2015).
Vardi, A. et al. Host–virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc. Natl Acad. Sci. USA 109, 19327–19332 (2012).
Møller, E. F. & Nielsen, T. G. Production of bacterial substrate by marine copepods: effect of phytoplankton biomass and cell size. J. Plankton Res. 23, 527–536 (2001).
Nagata, T. in Microbial Ecology of the Oceans (ed. Kirchman, D. L.) 121–152 (Wiley-Liss, 2000).
Strom, S. L., Benner, R., Ziegler, S. & Dagg, M. J. Planktonic grazers are a potentially important source of marine dissolved organic carbon. Limnol. Oceanogr. 42, 1364–1374 (1997).
Suttle, C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Zhao, Z. et al. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J. 13, 2551–2565 (2019).
Hurwitz, B. L. & U’Ren, J. M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol. 31, 161–168 (2016).
Kuhlisch, C. et al. Viral infection of algal blooms leaves a unique metabolic footprint on the dissolved organic matter in the ocean. Sci. Adv. 7, eabf4680 (2021).
Klawonn, I. et al. Characterizing the ‘fungal shunt’: parasitic fungi on diatoms affect carbon flow and bacterial communities in aquatic microbial food webs. Proc. Natl Acad. Sci. USA 118, e2102225118 (2021).
Agustí, S., Satta, M. P., Mura, M. P. & Benavent, E. Dissolved esterase activity as a tracer of phytoplankton lysis: evidence of high phytoplankton lysis rates in the northwestern Mediterranean. Limnol. Oceanogr. 43, 1836–1849 (1998).
Brussaard, C. et al. Effects of grazing, sedimentation and phytoplankton cell lysis on the structure of a coastal pelagic food web. Mar. Ecol. Prog. Ser. 123, 259–271 (1995).
Enke, T. N. et al. Modular Assembly of Polysaccharide-Degrading Marine Microbial Communities. Curr. Biol. 29, 1528–1535.e6 (2019).
Selander, E. et al. Predator lipids induce paralytic shellfish toxins in bloom-forming algae. Proc. Natl Acad. Sci. USA 112, 6395–6400 (2015).
Steinberg, D. K. & Landry, M. R. Zooplankton and the ocean carbon cycle. Ann. Rev. Mar. Sci. 9, 413–444 (2017).
Stoecker, D. K., Hansen, P. J., Caron, D. A. & Mitra, A. Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 9, 311–335 (2017).
Ankrah, N. Y. et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 8, 1089–1100 (2014).
Howard-Varona, C. et al. Phage-specific metabolic reprogramming of virocells. ISME J. 14, 881–895 (2020).
Saba, G. K., Steinberg, D. K. & Bronk, D. A. The relative importance of sloppy feeding, excretion, and fecal pellet leaching in the release of dissolved carbon and nitrogen by Acartia tonsa copepods. J. Exp. Mar. Biol. Ecol. 404, 47–56 (2011).
Bayer, B. et al. Ammonia‐oxidizing archaea release a suite of organic compounds potentially fueling prokaryotic heterotrophy in the ocean. Environ. Microbiol. 21, 4062–4075 (2019).
Kujawinski, E. B. et al. Identification of possible source markers in marine dissolved organic matter using ultrahigh resolution mass spectrometry. Geochim. Cosmochim. Acta 73, 4384–4399 (2009).
Schulz, S. & Dickschat, J. S. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842 (2007).
Wienhausen, G., Noriega-Ortega, B. E., Niggemann, J., Dittmar, T. & Simon, M. The exometabolome of two model strains of the Roseobacter group: a marketplace of microbial metabolites. Front. Microbiol. 8, 1985 (2017).
Ortega‐Retuerta, E. et al. Dissolved organic matter released by two marine heterotrophic bacterial strains and its bioavailability for natural prokaryotic communities. Environ. Microbiol. https://doi.org/10.1111/1462-2920.15306 (2020).
Kharbush, J. J. et al. Particulate organic carbon deconstructed: molecular and chemical composition of particulate organic carbon in the ocean. Front. Mar. Sci. 7, 518 (2020).
Enke, T. N., Leventhal, G. E., Metzger, M., Saavedra, J. T. & Cordero, O. X. Microscale ecology regulates particulate organic matter turnover in model marine microbial communities. Nat. Commun. 9, 2743 (2018).
Reintjes, G., Arnosti, C., Fuchs, B. & Amann, R. Selfish, sharing and scavenging bacteria in the Atlantic Ocean: a biogeographical study of bacterial substrate utilisation. ISME J. 13, 1119–1132 (2019).
Smith, D. C., Simon, M., Alldredge, A. L. & Azam, F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359, 139–142 (1992).
Thor, P., Dam, H. G. & Rogers, D. R. Fate of organic carbon released from decomposing copepod fecal pellets in relation to bacterial production and ectoenzymatic activity. Aquat. Microb. Ecol. 33, 279–288 (2003).
Cole, J. J., Findlay, S. & Pace, M. L. Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43, 1–10 (1988).
Azam, F. Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696 (1998).
Moran, M. A. et al. The ocean’s labile DOC supply chain. Limnol. Oceanogr. https://doi.org/10.1002/lno.12053 (2022).
Lindeman, R. L. The trophic-dynamic aspect of ecology. Ecology 23, 399–417 (1942).
Hollibaugh, J., Carruthers, A., Fuhrman, J. & Azam, F. Cycling of organic nitrogen in marine plankton communities studied in enclosed water columns. Mar. Biol. 59, 15–21 (1980).
Keil, R. G. & Kirchman, D. L. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquat. Microb. Ecol. 18, 293–300 (1999).
Boysen, A. K. et al. Particulate metabolites and transcripts reflect diel oscillations of microbial activity in the surface ocean. mSystems 6, e00896-20 (2021).
Dawson, H. M. et al. Potential of temperature-and salinity-driven shifts in diatom compatible solute concentrations to impact biogeochemical cycling within sea ice. Elementa https://doi.org/10.1525/elementa.421 (2020).
Durham, B. P. Deciphering metabolic currencies that support marine microbial networks. mSystems 6, e0076321 (2021).
Fiore, C. L., Longnecker, K., Kido Soule, M. C. & Kujawinski, E. B. Release of ecologically relevant metabolites by the cyanobacterium Synechococcus elongatus CCMP 1631. Environ. Microbiol. 17, 3949–3963 (2015).
Durham, B. P. et al. Recognition cascade and metabolite transfer in a marine bacteria‐phytoplankton model system. Environ. Microbiol. 19, 3500–3513 (2017).
Durham, B. P. et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl Acad. Sci. USA 112, 453–457 (2015).
Ferrer-González, F. X. et al. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 15, 762–773 (2020).
Gebser, B., Thume, K., Steinke, M. & Pohnert, G. Phytoplankton‐derived zwitterionic gonyol and dimethylsulfonioacetate interfere with microbial dimethylsulfoniopropionate sulfur cycling. MicrobiologyOpen 9, e1014 (2020).
Heal, K. R. et al. Marine community metabolomes carry fingerprints of phytoplankton community composition. mSystems 6, e01334-20 (2020).
Kiene, R. P., Linn, L. J. & Bruton, J. A. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–224 (2000).
Landa, M., Burns, A. S., Roth, S. J. & Moran, M. A. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677 (2017).
Shibl, A. A. et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl Acad. Sci. USA 117, 27445–27455 (2020).
Uchimiya, M., Schroer, W., Olofsson, M., Edison, A. S. & Moran, M. A. Diel investments in metabolite production and consumption in a model microbial system. ISME J. https://doi.org/10.1038/s41396-021-01172-w (2021).
Mopper, K. & Lindroth, P. Diel and depth variations in dissolved free amino acids and ammonium in the Baltic Sea determined by shipboard HPLC analysis 1. Limnol. Oceanogr. 27, 336–347 (1982).
Pomeroy, L. R., Macko, S., Ostrom, P. & Dunphy, J. The microbial food web in Arctic seawater: concentration of dissolved free amino acids and bacterial abundance and activity in the Arctic Ocean and in Resolute Passage. Mar. Ecol. Prog. Ser. 61, 31–40 (1990).
Liu, Q., Lu, X. X., Tolar, B. B., Mou, X. Z. & Hollibaugh, J. T. Concentrations, turnover rates and fluxes of polyamines in coastal waters of the South Atlantic Bight. Biogeochemistry 123, 117–133 (2015).
Nishibori, N., Yuasa, A., Sakai, M., Fujihara, S. & Nishio, S. Free polyamine concentrations in coastal seawater during phytoplankton bloom. Fish. Sci. 67, 79–83 (2001).
Benner, R. & Kaiser, K. Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 48, 118–128 (2003).
Borch, N. H. & Kirchmann, D. L. Concentration and composition of dissolved combined neutral sugars (polysaccharides) in seawater determined by HPLC-PAD. Mar. Chem. 57, 85–95 (1997).
Ittekkot, V., Brockmann, U., Michaelis, W. & Degens, E. T. Dissolved free and combined carbohydrates during a phytoplankton bloom in the northern North Sea. Mar. Ecol. Prog. Ser. 4, 299–305 (1981).
Lau, W. W., Keil, R. G. & Armbrust, E. V. Succession and diel transcriptional response of the glycolate-utilizing component of the bacterial community during a spring phytoplankton bloom. Appl. Environ. Microbiol. 73, 2440–2450 (2007).
Bertrand, E. M. & Allen, A. E. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front. Microbiol. 3, 375 (2012).
Gómez‐Consarnau, L. et al. Mosaic patterns of B‐vitamin synthesis and utilization in a natural marine microbial community. Environ. Microbiol. 20, 2809–2823 (2018).
Sanudo-Wilhelmy, S. A. et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc. Natl Acad. Sci. USA 109, 14041–14045 (2012).
Amin, S. A. et al. Photolysis of iron-siderophore chelates promotes bacterial–algal mutualism. Proc. Natl Acad. Sci. USA 106, 17071–17076 (2009).
D’Onofrio, A. et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264 (2010).
Kenney, G. E. et al. The biosynthesis of methanobactin. Science 359, 1411–1416 (2018).
Lhospice, S. et al. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 7, 17132 (2017).
Button, D., Robertson, B., Gustafson, E. & Zhao, X. Experimental and theoretical bases of specific affinity, a cytoarchitecture-based formulation of nutrient collection proposed to supercede the Michaelis–Menten paradigm of microbial kinetics. Appl. Environ. Microbiol. 70, 5511–5521 (2004).
Button, D., Robertson, B. R., Lepp, P. W. & Schmidt, T. M. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64, 4467–4476 (1998).
Heal, K. R. et al. Determination of four forms of vitamin B12 and other B vitamins in seawater by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 28, 2398–2404 (2014).
Mawji, E. et al. Hydroxamate siderophores: occurrence and importance in the Atlantic Ocean. Environ. Sci. Technol. 42, 8675–8680 (2008).
Morris, J. J., Lenski, R. E. & Zinser, E. R. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio https://doi.org/10.1128/mBio.00036-12 (2012)
Kazamia, E., Helliwell, K. E., Purton, S. & Smith, A. G. How mutualisms arise in phytoplankton communities: building eco‐evolutionary principles for aquatic microbes. Ecol. Lett. 19, 810–822 (2016).
Pohnert, G., Steinke, M. & Tollrian, R. Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol. Evol. 22, 198–204 (2007).
Vos, M. et al. Infochemicals structure marine, terrestrial and freshwater food webs: implications for ecological informatics. Ecol. Inf. 1, 23–32 (2006).
Amin, S. et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015).
Hmelo, L. & Van Mooy, B. A. Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 54, 127–133 (2009).
Schwartz, E. R., Poulin, R. X., Mojib, N. & Kubanek, J. Chemical ecology of marine plankton. Nat. Prod. Rep. 33, 843–860 (2016).
Nelson, C. E. & Wear, E. K. Microbial diversity and the lability of dissolved organic carbon. Proc. Natl Acad. Sci. USA 111, 7166–7167 (2014).
Amaral-Zettler, L. et al. in Life in the World’s Oceans: Diversity, Distribution and Abundance (ed. McIntyre, A.) 223–245 (Wiley, 2010).
De Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
Hertkorn, N. et al. Natural organic matter and the event horizon of mass spectrometry. Anal. Chem. 80, 8908–8919 (2008).
Lechtenfeld, O. J., Hertkorn, N., Shen, Y., Witt, M. & Benner, R. Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 6, 6711 (2015).
Longnecker, K., Sievert, S. M., Sylva, S. P., Seewald, J. S. & Kujawinski, E. B. Dissolved organic carbon compounds in deep-sea hydrothermal vent fluids from the East Pacific Rise at 9° 50′ N. Org. Geochem. 125, 41–49 (2018).
Romano, S. et al. Exo-metabolome of Pseudovibrio sp. FO-BEG1 analyzed by ultra-high resolution mass spectrometry and the effect of phosphate limitation. PLoS ONE 9, e96038 (2014).
Förster, J., Famili, I., Fu, P., Palsson, B. Ø. & Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13, 244–253 (2003).
Pacheco, A. R., Moel, M. & Segrè, D. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103 (2019).
Hertkorn, N. et al. Characterization of a major refractory component of marine dissolved organic matter. Geochim. Cosmochim. Acta 70, 2990–3010 (2006).
Steen, A. D. et al. Analytical and computational advances, opportunities, and challenges in marine organic biogeochemistry in an era of ‘omics’. Front. Mar. Sci. https://doi.org/10.3389/fmars.2020.00718 (2020).
Becker, J. W. et al. Closely related phytoplankton species produce similar suites of dissolved organic matter. Front. Microbiol. 5, 111 (2014).
Bouslimani, A., Sanchez, L. M., Garg, N. & Dorrestein, P. C. Mass spectrometry of natural products: current, emerging and future technologies. Nat. Prod. Rep. 31, 718–729 (2014).
Johnson, W. M. et al. Metabolite composition of sinking particles differs from surface suspended particles across a latitudinal transect in the South Atlantic. Limnol. Oceanogr. 65, 111–127 (2020).
Metwally, H., McAllister, R. G. & Konermann, L. Exploring the mechanism of salt-induced signal suppression in protein electrospray mass spectrometry using experiments and molecular dynamics simulations. Anal. Chem. 87, 2434–2442 (2015).
Kovacs, H., Moskau, D. & Spraul, M. Cryogenically cooled probes—a leap in NMR technology. Prog. Nucl. Magn. Reson. Spectrosc. 2, 131–155 (2005).
Aluwihare, L. I., Repeta, D. J. & Chen, R. F. A major biopolymeric component to dissolved organic carbon in surface sea water. Nature 387, 166–169 (1997).
Koprivnjak, J.-F. et al. Chemical and spectroscopic characterization of marine dissolved organic matter isolated using coupled reverse osmosis–electrodialysis. Geochim. Cosmochim. Acta 73, 4215–4231 (2009).
Dittmar, T., Koch, B., Hertkorn, N. & Kattner, G. A simple and efficient method for the solid‐phase extraction of dissolved organic matter (SPE‐DOM) from seawater. Limnol. Oceanogr. Meth. 6, 230–235 (2008).
Johnson, W. M., Soule, M. C. K. & Kujawinski, E. B. Extraction efficiency and quantification of dissolved metabolites in targeted marine metabolomics. Limnol. Oceanogr. Meth. 15, 417–428 (2017).
Lee, C. & Bada, J. L. Amino acids in equatorial Pacific Ocean water. Earth Planet Sci. Lett. 26, 61–68 (1975).
Yamashita, Y. & Tanoue, E. Chemical characteristics of amino acid-containing dissolved organic matter in seawater. Org. Geochem. 35, 679–692 (2004).
Zubkov, M. V., Tarran, G. A., Mary, I. & Fuchs, B. M. Differential microbial uptake of dissolved amino acids and amino sugars in surface waters of the Atlantic Ocean. J. Plankton Res. 30, 211–220 (2008).
Unanue, M. et al. Ectoenzymatic activity and uptake of monomers in marine bacterioplankton described by a biphasic kinetic model. Microb. Ecol. 37, 36–48 (1999).
Nissen, H., Nissen, P. & Azam, F. Multiphasic uptake of d-glucose by an oligotrophic marine bacterium. Mar. Ecol. Prog. Ser. 16, 155–160 (1984).
Pedler, B. E., Aluwihare, L. I. & Azam, F. Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl Acad. Sci. USA 111, 7202–7207 (2014).
Giovannoni, S. J., Thrash, J. C. & Temperton, B. Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565 (2014).
Norris, N., Levine, N. M., Fernandez, V. I. & Stocker, R. Mechanistic model of nutrient uptake explains dichotomy between marine oligotrophic and copiotrophic bacteria. PLoS Comp. Biol. 17, e1009023 (2021).
Stocker, R., Seymour, J. R., Samadani, A., Hunt, D. E. & Polz, M. F. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl Acad. Sci. USA 105, 4209–4214 (2008).
Keil, R. G. & Kirchman, D. L. Dissolved combined amino acids—chemical form and utilization by marine bacteria. Limnol. Oceanogr. 38, 1256–1270 (1993).
Mopper, K. et al. Determination of sugars in unconcentrated seawater and other natural waters by liquid chromatography and pulsed amperometric detection. Environ. Sci. Technol. 26, 133–138 (1992).
Zakem, E. J., Cael, B. & Levine, N. M. A unified theory for organic matter accumulation. Proc. Natl Acad. Sci. USA 118, e2016896118 (2021).
Moran, M. A., Reisch, C. R., Kiene, R. P. & Whitman, W. B. Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 4, 523–542 (2012).
Kiene, R. P. & Slezak, D. Low dissolved DMSP concentrations in seawater revealed by small‐volume gravity filtration and dialysis sampling. Limnol. Oceanogr. Meth. 4, 80–95 (2006).
Smriga, S., Fernandez, V. I., Mitchell, J. G. & Stocker, R. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc. Natl Acad. Sci. USA 113, 1576–1581 (2016).
Azam, F. & Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007).
Seymour, J. R., Amin, S. A., Raina, J.-B. & Stocker, R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2, 17065 (2017).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Fenn, J. B., Mann, M., Meng, C. K., Wong, S. F. & Whitehouse, C. M. Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 (1989).
Marshall, A. G., Hendrickson, C. L. & Jackson, G. S. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 17, 1–35 (1998).
Zubarev, R. A. & Makarov, A. Orbitrap mass spectrometry. Anal. Chem. 85, 5288–5296 (1998).
Longnecker, K. & Kujawinski, E. B. Mining mass spectrometry data: using new computational tools to find novel organic compounds in complex environmental mixtures. Org. Geochem. 110, 92–99 (2017).
Ardenkjaer-Larsen, J. H. et al. Facing and overcoming sensitivity challenges in biomolecular NMR spectroscopy. Angew. Chem. Int. Ed. 54, 9162–9185 (2015).
Ramaswamy, V. et al. Microsample cryogenic probes: technology and applications. eMagRes https://doi.org/10.1002/9780470034590.emrstm1315 (2007)
Edison, A. S. et al. NMR: unique strengths that enhance modern metabolomics research. Anal. Chem. 93, 478–499 (2020).
Anaraki, M. T. et al. NMR assignment of the in vivo Daphnia magna metabolome. Analyst 145, 5787–5800 (2020).
Qi, Y. et al. Absorption-mode: the next generation of Fourier transform mass spectra. Anal. Chem. 84, 2923–2929 (2012).
Liaghati Mobarhan, Y., Soong, R., Lane, D. & Simpson, A. J. In vivo comprehensive multiphase NMR. Mag. Res. Chem. 58, 427–444 (2020).
Zhao, S., Li, H., Han, W., Chan, W. & Li, L. Metabolomic coverage of chemical-group-submetabolome analysis: group classification and four-channel chemical isotope labeling LC–MS. Anal. Chem. 91, 12108–12115 (2019).
Sogin, E. M., Puskas, E., Dubilier, N. & Liebeke, M. Marine metabolomics: measurement of metabolites in seawater by gas chromatography mass spectrometry. mSystems 4, e00638-19 (2019).
Pontrelli, S. & Sauer, U. Salt-tolerant metabolomics for exometabolomic measurements of marine bacterial isolates. Anal. Chem. 93, 7164–7171 (2021).
Mayali, X., Weber, P. K., Mabery, S. & Pett-Ridge, J. Phylogenetic patterns in the microbial response to resource availability: amino acid incorporation in San Francisco Bay. PLoS ONE 9, e95842 (2014).
Thomas, F. et al. Isotopic tracing reveals single-cell assimilation of a macroalgal polysaccharide by a few marine Flavobacteria and Gammaproteobacteria. ISME J. 15, 3062–3075 (2021).
Hatzenpichler, R. et al. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ. Microbiol. 16, 2568–2590 (2014).
Martinez-Garcia, M. et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS ONE 7, e35314 (2012).
Raina, J.-B. et al. Subcellular tracking reveals the location of dimethylsulfoniopropionate in microalgae and visualises its uptake by marine bacteria. eLife 6, e23008 (2017).
Vasdekis, A. E. & Stephanopoulos, G. Review of methods to probe single cell metabolism and bioenergetics. Metab. Eng. 27, 115–135 (2015).
Baumeister, T., Vallet, M., Kaftan, F., Svatoš, A. & Pohnert, G. Live single-cell metabolomics with matrix-free laser/desorption ionization mass spectrometry to address microalgal physiology. Front. Plant Sci. 10, 172 (2019).
Schleyer, G. et al. In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids. Nat. Microbiol. 4, 527–538 (2019).
Judge, M. T. et al. Continuous in vivo metabolism by NMR. Front. Mol. Biosci. 6, 26 (2019).
Gao, C. et al. Single-cell bacterial transcription measurements reveal the importance of dimethylsulfoniopropionate (DMSP) hotspots in ocean sulfur cycling. Nat. Commun. 11, 1942 (2020).
Szul, M. J., Dearth, S. P., Campagna, S. R. & Zinser, E. R. Carbon fate and flux in Prochlorococcus under nitrogen limitation. mSystems 4, e00254-18 (2019).
Teeling, H. et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 (2012).
Nowinski, B. & Moran, M. A. Niche dimensions of a marine bacterium are identified using invasion studies in coastal seawater. Nat. Microbiol. 6, 524–532 (2021).
Poretsky, R. S. et al. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific Subtropical Gyre. Environ. Microbiol. 11, 1358–1375 (2009).
Sowell, S. M. et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3, 93–105 (2009).
Lidbury, I., Murrell, J. C. & Chen, Y. Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc. Natl Acad. Sci. USA 111, 2710–2715 (2014).
Tang, K., Jiao, N., Liu, K., Zhang, Y. & Li, S. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS ONE 7, e41204 (2012).
Lima-Mendez, G. et al. Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015).
Reverter, M., Rohde, S., Parchemin, C., Tapissier-Bontemps, N. & Schupp, P. J. Metabolomics and marine biotechnology: coupling metabolite profiling and organism biology for the discovery of new compounds. Front. Mar. Sci. 7, 1062 (2020).
Frischkorn, K. R., Rouco, M., Van Mooy, B. A. & Dyhrman, S. T. Epibionts dominate metabolic functional potential of Trichodesmium colonies from the oligotrophic ocean. ISME J. 11, 2090–2101 (2017).
Vorobev, A. et al. Identifying labile DOM components in a coastal ocean through depleted bacterial transcripts and chemical signals. Environ. Microbiol. 20, 3012–3030 (2018).
Becker, S. et al. Laminarin is a major molecule in the marine carbon cycle. Proc. Natl Acad. Sci. USA 117, 6599–6607 (2020).
Glavinas, H. et al. Utilization of membrane vesicle preparations to study drug–ABC transporter interactions. Expert Opin. Drug Metab. Toxicol. 4, 721–732 (2008).
Piazza, I. et al. A map of protein–metabolite interactions reveals principles of chemical communication. Cell 172, 358–372.e23 (2018).
Chauvigné-Hines, L. M. et al. Suite of activity-based probes for cellulose-degrading enzymes. J. Am. Chem. Soc. 134, 20521–20532 (2012).
Baklouti, M., Diaz, F., Pinazo, C., Faure, V. & Quéguiner, B. Investigation of mechanistic formulations depicting phytoplankton dynamics for models of marine pelagic ecosystems and description of a new model. Prog. Oceanogr. 71, 1–33 (2006).
Kovač, Ž., Platt, T., Sathyendranath, S. & Lomas, M. Extraction of photosynthesis parameters from time series measurements of in situ production: Bermuda Atlantic Time-Series Study. Remote Sens. 10, 915 (2018).
Mongin, M., Nelson, D. M., Pondaven, P., Brzezinski, M. A. & Tréguer, P. Simulation of upper-ocean biogeochemistry with a flexible-composition phytoplankton model: C, N and Si cycling in the western Sargasso Sea. Deep Sea Res. I 50, 1445–1480 (2003).
Karr, J. R. et al. A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 (2012).
Malik, A. A. et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14, 1–9 (2020).
Ofaim, S., Sulheim, S., Almaas, E., Sher, D. & Segrè, D. Dynamic allocation of carbon storage and nutrient-dependent exudation in a revised genome-scale model of Prochlorococcus. Front. Genet. 12, 91 (2021).
Colarusso, A. V., Goodchild-Michelman, I., Rayle, M. & Zomorrodi, A. R. Computational modeling of metabolism in microbial communities on a genome-scale. Curr. Opin. Syst. Biol. https://doi.org/10.1016/j.coisb.2021.04.001 (2021).
Harvey, E. L. et al. A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore, Emiliania huxleyi. Front. Microbiol. 7, 59 (2016).
Skalnik, C. J. et al. Whole-colony modeling of Escherichia coli. Preprint at bioRxiv https://doi.org/10.1101/2021.04.27.441666 (2021).
Reid, A. Incorporating Microbial Processes into Climate Models (American Academy of Microbiology, 2012).
Nicholson, D. P., Stanley, R. H. & Doney, S. C. A phytoplankton model for the allocation of gross photosynthetic energy including the trade‐offs of diazotrophy. J. Geophys. Res. Biogeosci. 123, 1796–1816 (2018).
Bonan, G. B. & Doney, S. C. Climate, ecosystems, and planetary futures: the challenge to predict life in Earth system models. Science 359, eaam8328 (2018).
Coles, V. et al. Ocean biogeochemistry modeled with emergent trait-based genomics. Science 358, 1149–1154 (2017).
Follows, M. J., Dutkiewicz, S., Grant, S. & Chisholm, S. W. Emergent biogeography of microbial communities in a model ocean. Science 315, 1843–1846 (2007).
Lane, N. & Martin, W. The energetics of genome complexity. Nature 467, 929–934 (2010).
Lynch, M. & Marinov, G. K. The bioenergetic costs of a gene. Proc. Natl Acad. Sci. USA 112, 15690–15695 (2015).
Bolduc, B., Youens-Clark, K., Roux, S., Hurwitz, B. L. & Sullivan, M. B. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 11, 7–14 (2017).
Eren, A. M. et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 6, 3–6 (2021).
Harcombe, W. R. et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115 (2014).
Saito, M. A. et al. Development of an Ocean Protein Portal for interactive discovery and education. J. Proteome Res. 20, 326–336 (2020).
Durán, C. et al. Nonlinear machine learning pattern recognition and bacteria-metabolite multilayer network analysis of perturbed gastric microbiome. Nat. Commun. 12, 1926 (2021).
Erbilgin, O. et al. MAGI: a method for metabolite annotation and gene integration. ACS Chem. Biol. 14, 704–714 (2019).
Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016).
Schorn, M. A. et al. A community resource for paired genomic and metabolomic data mining. Nat. Chem. Biol. 17, 363–368 (2021).
Goldford, J. E. et al. Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018).
Zomorrodi, A. R. & Segrè, D. Genome-driven evolutionary game theory helps understand the rise of metabolic interdependencies in microbial communities. Nat. Commun. 8, 1563 (2017).
Pachiadaki, M. G. et al. Charting the complexity of the marine microbiome through single-cell genomics. Cell 179, 1623–1635.e11 (2019).
Paoli, L. et al. Uncharted biosynthetic potential of the ocean microbiome. Preprint at bioRxiv https://doi.org/10.1101/2021.03.24.436479 (2021).
Delmont, T. O. et al. Functional repertoire convergence of distantly related eukaryotic plankton lineages revealed by genome-resolved metagenomics. Preprint at bioRxiv https://doi.org/10.1101/2020.10.15.341214 (2021).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Acinas, S. G. et al. Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun. Biol. 4, 604 (2021).
Salazar, G. et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179, 1068–1083.e21 (2019).
Saito, M. A. et al. Progress and challenges in ocean metaproteomics and proposed best practices for data sharing. J. Proteome Res. 18, 1461–1476 (2019).
Chandler, C. et al. Effective management of ocean biogeochemistry and ecological data: the BCO-DMO story. In EGU General Assembly Conference Abstracts Vol. 14, 1258 (2012).
Ducklow, H. W., Steinberg, D. K. & Buesseler, K. O. Upper ocean carbon export and the biological pump. Oceanography 14, 50–58 (2001).
Siegel, D. A., DeVries, T., Doney, S. C. & Bell, T. Assessing the sequestration time scales of some ocean-based carbon dioxide reduction strategies. Environ. Res. Lett. 16, 104003 (2021).
Del Giorgio, P. A. & Duarte, C. M. Respiration in the open ocean. Nature 420, 379–384 (2002).
Rivkin, R. B. & Legendre, L. Biogenic carbon cycling in the upper ocean: effects of microbial respiration. Science 291, 2398–2400 (2001).
Toseland, A. et al. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Change 3, 979–984 (2013).
Brembu, T., Mühlroth, A., Alipanah, L. & Bones, A. M. The effects of phosphorus limitation on carbon metabolism in diatoms. Phil. Trans. R. Soc. B 372, 20160406 (2017).
Liang, Y., Koester, J. A., Liefer, J. D., Irwin, A. J. & Finkel, Z. V. Molecular mechanisms of temperature acclimation and adaptation in marine diatoms. ISME J. 13, 2415–2425 (2019).
Hennon, G. M., Hernández Limón, M. D., Haley, S. T., Juhl, A. R. & Dyhrman, S. T. Diverse CO2-induced responses in physiology and gene expression among eukaryotic phytoplankton. Front. Microbiol. 8, 2547 (2017).
Shi, D. et al. Interactive effects of light, nitrogen source, and carbon dioxide on energy metabolism in the diatom Thalassiosira pseudonana. Limnol. Oceanogr. 60, 1805–1822 (2015).
Taylor, A. G. et al. Sharp gradients in phytoplankton community structure across a frontal zone in the California Current ecosystem. J. Plankton Res. 34, 778–789 (2012).
Marinov, I., Doney, S. & Lima, I. Response of ocean phytoplankton community structure to climate change over the 21st century: partitioning the effects of nutrients, temperature and light. Biogeosciences 7, 3941–3959 (2010).
Becker, K. W. et al. Daily changes in phytoplankton lipidomes reveal mechanisms of energy storage in the open ocean. Nat. Commun. 9, 5179 (2018).
Johnson, W. M., Soule, M. C. K. & Kujawinski, E. B. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 10, 2304–2316 (2016).
Matrai, P. & Keller, M. Total organic sulfur and dimethylsulfoniopropionate in marine phytoplankton: intracellular variations. Mar. Biol. 119, 61–68 (1994).
Thume, K. et al. The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature 563, 412–415 (2018).
Baines, S. B. & Pace, M. L. The production of dissolved organic matter by phytoplankton and its importance to bacteria: patterns across marine and freshwater systems. Limnol. Oceanogr. 36, 1078–1090 (1991).
Peironcely, J. E., Reijmers, T., Coulier, L., Bender, A. & Hankemeier, T. Understanding and classifying metabolite space and metabolite-likeness. PLoS ONE 6, e28966 (2011).
Gifford, S. M., Sharma, S., Booth, M. & Moran, M. A. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 7, 281–298 (2013).
Poretsky, R. S., Sun, S., Mou, X. & Moran, M. A. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12, 616–627 (2010).
McCarren, J. et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl Acad. Sci. USA 107, 16420–16427 (2010).
Ziervogel, K. & Arnosti, C. Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate‐free seawater from the north‐eastern Gulf of Mexico. Environ. Microbiol. 10, 289–299 (2008).
Landa, M. et al. Sulfur metabolites that facilitate oceanic phytoplankton–bacteria carbon flux. ISME J. 13, 2536–2550 (2019).
Amin, S. A., Parker, M. S. & Armbrust, E. V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012).
Cude, W. N. et al. The production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization in the marine Roseobacter Phaeobacter sp. strain Y4I. Appl. Environ. Microbiol. 78, 4771–4780 (2012).
Edwards, B. R., Bidle, K. D. & Van Mooy, B. A. Dose-dependent regulation of microbial activity on sinking particles by polyunsaturated aldehydes: implications for the carbon cycle. Proc. Natl Acad. Sci. USA 112, 5909–5914 (2015).
Gram, L., Grossart, H.-P., Schlingloff, A. & Kiørboe, T. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68, 4111–4116 (2002).
Seyedsayamdost, M. R., Case, R. J., Kolter, R. & Clardy, J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335 (2011).
Teasdale, M. E., Liu, J., Wallace, J., Akhlaghi, F. & Rowley, D. C. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl. Environ. Microbiol. 75, 567–572 (2009).
Ross, A. C., Gulland, L. E., Dorrestein, P. C. & Moore, B. S. Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth. Biol. 4, 414–420 (2014).
Ziemert, N. et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl Acad. Sci. USA 111, E1130–E1139 (2014).
Kiene, R. P. & Linn, L. J. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45, 849–861 (2000).
Andreae, M. O. & Crutzen, P. J. Atmospheric aerosols: biogeochemical sources and role in atmospheric chemistry. Science 276, 1052–1058 (1997).
Charlson, R. J., Lovelock, J. E., Andreae, M. O. & Warren, S. G. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326, 655–661 (1987).
Curson, A. R. et al. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2, 17009 (2017).
Howard, E. C. et al. Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652 (2006).
Todd, J., Curson, A., Dupont, C., Nicholson, P. & Johnston, A. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ. Microbiol. 11, 1376–1385 (2009).
Todd, J. D. et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007).
Vila-Costa, M. et al. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314, 652–654 (2006).
Evans, C. et al. The relative significance of viral lysis and microzooplankton grazing as pathways of dimethylsulfoniopropionate (DMSP) cleavage: an Emiliania huxleyi culture study. Limnol. Oceanogr. 52, 1036–1045 (2007).
Bratbak, G. et al. Viral activity in relation to Emiliania huxleyi blooms: a mechanism of DMSP release? Mar. Ecol. Prog. Ser. 128, 133–142 (1995).
Seymour, J. R., Simó, R., Ahmed, T. & Stocker, R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345 (2010).
Barak-Gavish, N. et al. Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci. Adv. 4, eaau5716 (2018).
McParland, E. L. & Levine, N. M. The role of differential DMSP production and community composition in predicting variability of global surface DMSP concentrations. Limnol. Oceanogr. 64, 757–773 (2019).
Varaljay, V. A. et al. Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 9, 1677–1686 (2015).
Acknowledgements
This Review arose from discussions supported by the CIFAR and developed further with support from the Moore Foundation (5503 to M.A.M., E.B.K. and A.S.E.; 9335 to S.A.A.), the Simons Foundation (542391 to M.A.M.; 253417 to E.B.K.; 504183 to E.M.B.; 509034SCFY20 to R.B.; 721225 to S.T.D.; 687269 to A.M.E.; 542389 to N.M.L.; 345889 to A.E.S.; and 735079 to A.V.), the US NSF (OCE-1756105 to N.R.B.; Major Research Equipment DBI-1946970 to A.S.E.; OCE-1924554 to M.A.S.; CAREER DEB-1749544 to A.S.; OCE-BSF 1635070 to DS.; and OCE-1829831 to M.B.S.), the Paul M. Angell Family Foundation (to S.T.D.), and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-06159 to L.L.; RGPIN-2015-06078 to A.C.R.). This is NSF Center for Chemical Currencies of a Microbial Planet (C-CoMP) publication number 001.
Author information
Authors and Affiliations
Contributions
All authors contributed ideas. M.A.M., E.B.K. and W.F.S. wrote the manuscript with substantial input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Pieter Dorrestein, Manuel Liebeke and Bethanie Edwards for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Moran, M.A., Kujawinski, E.B., Schroer, W.F. et al. Microbial metabolites in the marine carbon cycle. Nat Microbiol 7, 508–523 (2022). https://doi.org/10.1038/s41564-022-01090-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01090-3
This article is cited by
-
Algal blooms in the ocean: hot spots for chemically mediated microbial interactions
Nature Reviews Microbiology (2024)
-
Jellyfish detritus supports niche partitioning and metabolic interactions among pelagic marine bacteria
Microbiome (2023)
-
Progress and challenges in exploring aquatic microbial communities using non-targeted metabolomics
The ISME Journal (2023)
-
Bacterial transcriptional response to labile exometabolites from photosynthetic picoeukaryote Micromonas commoda
ISME Communications (2023)
-
Plastoquinone synthesis inhibition by tetrabromo biphenyldiol as a widespread algicidal mechanism of marine bacteria
The ISME Journal (2023)