Abstract
Vaginal microbiota composition affects many facets of reproductive health. Lactobacillus iners-dominated microbial communities are associated with poorer outcomes, including higher risk of bacterial vaginosis (BV), compared with vaginal microbiota rich in L. crispatus. Unfortunately, standard-of-care metronidazole therapy for BV typically results in dominance of L. iners, probably contributing to post-treatment relapse. Here we generate an L. iners isolate collection comprising 34 previously unreported isolates from 14 South African women with and without BV and 4 previously unreported isolates from 3 US women. We also report an associated genome catalogue comprising 1,218 vaginal Lactobacillus isolate genomes and metagenome-assembled genomes from >300 women across 4 continents. We show that, unlike L. crispatus, L. iners growth is dependent on l-cysteine in vitro and we trace this phenotype to the absence of canonical cysteine biosynthesis pathways and a restricted repertoire of cysteine-related transport mechanisms. We further show that cysteine concentrations in cervicovaginal lavage samples correlate with Lactobacillus abundance in vivo and that cystine uptake inhibitors selectively inhibit L. iners growth in vitro. Combining an inhibitor with metronidazole promotes L. crispatus dominance of defined BV-like communities in vitro by suppressing L. iners growth. Our findings enable a better understanding of L. iners biology and suggest candidate treatments to modulate the vaginal microbiota to improve reproductive health for women globally.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Compressed directories containing data files sufficient to reproduce (1) analysis of pan-genome composition and gene content, (2) analysis for human FGT microbiota–metabolite analysis and (3) analysis for competition cultures of L. iners and L. crispatus and mixed community cultures are posted at Zenodo.org under https://zenodo.org/record/590046981. The dataset containing the raw Illumina MiSeq read data for genital tract bacterial 16S rRNA gene profiling analysed in this study (Fig. 3 and Extended Data Figs. 3 and 4) is available in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA729907. The dataset containing the raw Illumina MiSeq read data for the bacterial 16S rRNA gene sequences from competition cultures of L. iners and L. crispatus (Fig. 5a) and from mixed community cultures (Fig. 5c,d) is available in the NCBI SRA under BioProject PRJNA777644. The taxonomic assignments used for amplicon sequence variants (ASVs) from bacterial 16S rRNA gene sequencing are supplied in Supplementary Table 15. The Lactobacillus genomic catalogues included a total of 1,091 previously unreported isolate genomes, partial genomes and MAGs from multiple human cohorts, as detailed above. The assemblies are available in the NCBI SRA under BioProjects PRJNA799384, PRJNA799634, PRJNA799626, PRJNA799445, PRJNA799630, PRJNA799633, PRJNA799642, PRJNA799746, PRJNA799744, PRJNA799737, PRJNA799762 and PRJNA797778; additional details on the individual studies associated with these BioProjects are contained in Supplementary Tables 5, 6 and 10, and individual NCBI BioSample accession numbers for each of the 1,091 assemblies are listed in Supplementary Table 8. In addition, the genome catalogues included 127 previously reported isolate genomes that were retrieved from RefSeq; the individual accession numbers for these genomes are listed in Supplementary Table 7. The raw and corrected cystine and serine isotopologue measurements associated with Figs. 2c,d and 4c are available in Supplementary Tables 13 and 14. Some metadata related to previously reported isolate genomes were obtained from corresponding entries in RefSeq (https://www.ncbi.nlm.nih.gov/refseq/) or the Genomes OnLine Database (GOLD; https://gold.jgi.doe.gov/). Source data are supplied for plots and phylogenetic trees, including for Figs. 1–5 and Extended Data Figs. 1–8.
Code availability
Compressed directories containing R analysis code sufficient to reproduce (1) analysis of pan-genome composition and gene content, (2) analysis for human FGT microbiota–metabolite analysis and (3) analysis for competition cultures of L. iners and L. crispatus and mixed community cultures are available at Zenodo.org under https://zenodo.org/record/590046981. Each compressed directory contains a README file describing dependencies and other details, an R Project file, an R Markdown file containing the analysis code with additional information, and associated subdirectories used in the analysis.
References
Gosmann, C. et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 46, 29–37 (2017).
McClelland, R. S. et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect. Dis. 18, 554–564 (2018).
Fettweis, J. M. et al. The vaginal microbiome and preterm birth. Nat. Med. 25, 1012–1021 (2019).
Anahtar, M. N. et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42, 965–976 (2015).
Jespers, V. et al. A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa. Sci. Rep. 7, 11974 (2017).
Lennard, K. et al. Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect. Immun. 86, e00410–e00417 (2018).
Norenhag, J. et al. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG 127, 171–180 (2020).
Anahtar, M. N., Gootenberg, D. B., Mitchell, C. M. & Kwon, D. S. Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host Microbe 23, 159–168 (2018).
McKinnon, L. R. et al. The evolving facets of bacterial vaginosis: implications for HIV transmission. AIDS Res. Hum. Retroviruses 35, 219–228 (2019).
Kenyon, C., Colebunders, R. & Crucitti, T. The global epidemiology of bacterial vaginosis: a systematic review. Am. J. Obstet. Gynecol. 209, 505–523 (2013).
Vaneechoutte, M. Lactobacillus iners, the unusual suspect. Res. Microbiol. 168, 826–836 (2017).
Kindinger, L. M. et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome 5, 6 (2017).
Brooks, J. P. et al. Changes in vaginal community state types reflect major shifts in the microbiome. Microb. Ecol. Health Dis. 28, 1303265 (2017).
DiGiulio, D. B. et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl Acad. Sci. USA 112, 11060–11065 (2015).
Lambert, J. A., John, S., Sobel, J. D. & Akins, R. A. Longitudinal analysis of vaginal microbiome dynamics in women with recurrent bacterial vaginosis: recognition of the conversion process. PLoS ONE 8, e82599 (2013).
Munoz, A. et al. Modeling the temporal dynamics of cervicovaginal microbiota identifies targets that may promote reproductive health. Microbiome 9, 163 (2021).
Srinivasan, S. et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS ONE 5, e10197 (2010).
Petrova, M. I., Reid, G., Vaneechoutte, M. & Lebeer, S. Lactobacillus iners: friend or foe? Trends Microbiol. 25, 182–191 (2017).
Schwebke, J. R. et al. Treatment of male sexual partners of women with bacterial vaginosis: a randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 73, e672–e679 (2021).
Bradshaw, C. S. et al. High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence. J. Infect. Dis. 193, 1478–1486 (2006).
Joag, V. et al. Impact of standard bacterial vaginosis treatment on the genital microbiota, immune milieu, and ex vivo human immunodeficiency virus susceptibility. Clin. Infect. Dis. 68, 1675–1683 (2019).
Mitchell, C. et al. Behavioral predictors of colonization with Lactobacillus crispatus or Lactobacillus jensenii after treatment for bacterial vaginosis: a cohort study. Infect. Dis. Obstet. Gynecol. 2012, 706540 (2012).
Ravel, J. et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1, 29 (2013).
Verwijs, M. C., Agaba, S. K., Darby, A. C. & van de Wijgert, J. H. H. M. Impact of oral metronidazole treatment on the vaginal microbiota and correlates of treatment failure. Am. J. Obstet. Gynecol. 222, 157.e1–157.e13 (2020).
Cohen, C. R. et al. Randomized trial of lactin-V to prevent recurrence of bacterial vaginosis. N. Engl. J. Med. 382, 1906–1915 (2020).
Rampersaud, R. et al. Inerolysin, a cholesterol-dependent cytolysin produced by Lactobacillus iners. J. Bacteriol. 193, 1034–1041 (2011).
Antonio, M. A. D., Rabe, L. K. & Hillier, S. L. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J. Infect. Dis. 192, 394–398 (2005).
France, M. T., Mendes-Soares, H. & Forney, L. J. Genomic comparisons of Lactobacillus crispatus and Lactobacillus iners reveal potential ecological drivers of community composition in the vagina. Appl. Environ. Microbiol. 82, 7063–7073 (2016).
Witkin, S. S. et al. Influence of vaginal bacteria and d- and l-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections. mBio 4, e00460–13 (2013).
De Man, J. C., Rogosa, M. & Sharpe, M. E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23, 130–135 (1960).
Falsen, E., Pascual, C., Sjödén, B., Ohlén, M. & Collins, M. D. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int. J. Syst. Bacteriol. 49, 217–221 (1999).
Damelin, L. H. et al. Identification of predominant culturable vaginal Lactobacillus species and associated bacteriophages from women with and without vaginal discharge syndrome in South Africa. J. Med. Microbiol. 60, 180–183 (2011).
Manhanzva, M. T. et al. Inflammatory and antimicrobial properties differ between vaginal Lactobacillus isolates from South African women with non-optimal versus optimal microbiota. Sci. Rep. 10, 6196 (2020).
Matsumoto, A. et al. Characterization of the vaginal microbiota of Japanese women. Anaerobe 54, 172–177 (2018).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Guédon, E. & Martin-Verstraete, I. in Amino Acid Biosynthesis ~ Pathways, Regulation and Metabolic Engineering (ed. Wendisch, V. F.) 195–218 (Springer, 2006); https://doi.org/10.1007/7171_2006_060
Dong, K. L. et al. Detection and treatment of Fiebig stage I HIV-1 infection in young at-risk women in South Africa: a prospective cohort study. Lancet HIV 5, e35–e44 (2018).
Nugent, R. P., Krohn, M. A. & Hillier, S. L. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J. Clin. Microbiol. 29, 297–301 (1991).
McIver, C. J. & Tapsall, J. W. Cysteine requirements of naturally occurring cysteine auxotrophs of Escherichia coli. Pathology 19, 361–363 (1987).
Proust, L. et al. Insights into the complexity of yeast extract peptides and their utilization by Streptococcus thermophilus. Front. Microbiol. 10, 906 (2019).
Burguière, P., Auger, S., Hullo, M.-F., Danchin, A. & Martin-Verstraete, I. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186, 4875–4884 (2004).
Müller, A. et al. An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol. Microbiol. 57, 143–155 (2005).
Zhou, Y. & Imlay, J. A. Escherichia coli k-12 lacks a high-affinity assimilatory cysteine importer. mBio 11, e01073–20 (2020).
Poole, R. K., Cozens, A. G. & Shepherd, M. The CydDC family of transporters. Res. Microbiol. 170, 407–416 (2019).
Pophaly, S. D., Singh, R., Pophaly, S. D., Kaushik, J. K. & Tomar, S. K. Current status and emerging role of glutathione in food grade lactic acid bacteria. Microb. Cell Fact. 11, 114 (2012).
Suzuki, H., Koyanagi, T., Izuka, S., Onishi, A. & Kumagai, H. The yliA, -B, -C, and -D genes of Escherichia coli K-12 encode a novel glutathione importer with an ATP-binding cassette. J. Bacteriol. 187, 5861–5867 (2005).
Rampersaud, R. Identification and Characterization of Inerolysin, the Cholesterol Dependent Cytolysin produced by Lactobacillus iners (Columbia University, 2014).
Klatt, N. R. et al. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356, 938–945 (2017).
Borgdorff, H. et al. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS ONE 12, e0181135 (2017).
Borgdorff, H. et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 8, 1781–1793 (2014).
Chen, Y. et al. Association between the vaginal microbiome and high-risk human papillomavirus infection in pregnant Chinese women. BMC Infect. Dis. 19, 677 (2019).
Marconi, C. et al. Characterization of the vaginal microbiome in women of reproductive age from 5 regions in Brazil. Sex. Transm. Dis. 47, 562–569 (2020).
Plummer, E. L. et al. Sexual practices have a significant impact on the vaginal microbiota of women who have sex with women. Sci. Rep. 9, 19749 (2019).
Vargas-Robles, D. et al. Changes in the vaginal microbiota across a gradient of urbanization. Sci. Rep. 10, 12487 (2020).
Mendes-Soares, H., Suzuki, H., Hickey, R. J. & Forney, L. J. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J. Bacteriol. 196, 1458–1470 (2014).
Srinivasan, S. et al. Metabolic signatures of bacterial vaginosis. mBio 6, e00204–e00215 (2015).
Castellano, F., Correale, J. & Molinier-Frenkel, V. Editorial: immunosuppressive amino acid catabolizing enzymes in heallth and disease. Front. Immunol. 12, 689864 (2021).
Lev-Sagie, A. et al. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat. Med. 25, 1500–1504 (2019).
Fettweis, J. M. et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 160, 2272–2282 (2014).
Hoang, T. et al. The cervicovaginal mucus barrier to HIV-1 is diminished in bacterial vaginosis. PLoS Pathog. 16, e1008236 (2020).
Vitali, B. et al. Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2367–2376 (2015).
McMillan, A. et al. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci. Rep. 5, 14174 (2015).
Callahan, B. J. et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl Acad. Sci. USA 114, 9966–9971 (2017).
Marrazzo, J. M., Thomas, K. K., Fiedler, T. L., Ringwood, K. & Fredricks, D. N. Relationship of specific vaginal bacteria and bacterial vaginosis treatment failure in women who have sex with women: a cohort study. Ann. Intern. Med. 149, 20 (2008).
Amsel, R. et al. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74, 14–22 (1983).
Mitchell, C. M. et al. Vaginal microbiota and mucosal immune markers in women with vulvovaginal discomfort. Sex. Transm. Dis. 47, 269–274 (2020).
Methé, B. A. et al. A framework for human microbiome research. Nature 486, 215–221 (2012).
Consortium, H. M. P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Goltsman, D. S. A. et al. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 28, 1467–1480 (2018).
Hudson, P. L. et al. Comparison of the vaginal microbiota in postmenopausal black and white women. J. Infect. Dis. https://doi.org/10.1093/infdis/jiaa780 (2020).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145.e5 (2018).
Li, F. et al. The metagenome of the female upper reproductive tract. Gigascience 7, giy107 (2018).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522 (2011).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Rocha, J. et al. Lactobacillus mulieris sp. nov., a new species of Lactobacillus delbrueckii group. Int. J. Syst. Evol. Microbiol. 70, 1522–1527 (2020).
France, M. T. et al. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 8, 166 (2020).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Bloom, S. M. & Kwon, D. S. Cysteine Dependence of Lactobacillus iners is a Potential Therapeutic Target for Vaginal Microbiota Modulation (Zenodo, 2021); https://doi.org/10.5281/ZENODO.5900469
Huerta-Cepas, J. et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Milanese, A. et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 10, 1014 (2019).
Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
McMurdie, P. J. & Holmes, S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Sutton, T. R. et al. A robust and versatile mass spectrometry platform for comprehensive assessment of the thiol redox metabolome. Redox Biol. 16, 359–380 (2018).
Hill, J. W., Coy, R. B. & Lewandowski, P. E. Oxidation of cysteine to cystine using hydrogen peroxide. J. Chem. Educ. 67, 172 (1990).
Millard, P. et al. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 35, 4484–4487 (2019).
Acknowledgements
We thank study participants for donating clinical samples used in this study; study staff at the FRESH cohort; laboratory staff at the HIV Pathogenesis Programme at UKZN for sample processing; J. A. Elsherbini (Ragon Institute) for bioinformatic support; L. Froehle (Ragon Institute) for helpful discussions of analysis; and D. Jenkins (Harvard Department of Chemistry and Chemical Biology) and M. Farcasanu, K. Jackson, L. Froehle and J. Bramante (Ragon Institute) for assay and sample assistance. J. Ravel and M. France (University of Maryland) and the Vaginal Microbiome Research Consortium (VMRC) kindly provided 4 experimental isolates (details in Supplementary Table 3, referred to as ‘VMRC’) as well as WGS data from 34 study participants and 312 previously unreported isolate genomes (details summarized in Supplementary Tables 5, 6, 8, 9 and 11; referred to as ‘VMRC’). J. Marrazzo (University of Alabama, Birmingham) kindly provided 111 strains of non-iners lactobacilli that were sequenced for genomic analysis (details summarized in Supplementary Tables 5, 6, 8, 9 and 11; referred to as ‘Vaginal Health Project’). This work was supported in part by National Institutes of Health grant NIH 1R01AI111918-01 to D.S.K. and by NIH grant T32 AI007387 to S.M.B.; additional NIH support was provided by grants to S.M.B. and M.S.G. from the Harvard University Center for AIDS Research (CFAR), an NIH funded programme (P30 AI060354) supported by the following NIH co-funding and participating institutes and centres: NIAID, NCI, NICHD, NIDCR, NHLBI, NIDA, NIMH, NIA, NIDDK, NINR, NIMHD, FIC and OAR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The work was also supported in part by: Bill and Melinda Gates Foundation grants OPP1189208 to D.S.K. and OPP1158186 to E.P.B. and D.A.R.; a Burroughs Wellcome Career Award for Medical Scientists to D.S.K.; Vincent Memorial Research Funds and a Domolky Innovation Award (Massachusetts General Hospital) to C.M.M.; the South African Research Chairs Initiative through the National Research Foundation and the Victor Daitz Foundation to T.N.; funds from the Thomas C. and Joan M. Merigan Endowment at Stanford to D.A.R.; funding from the Harvard Program for Research in Science and Engineering (PRISE) and the Harvard Microbial Sciences Initiative to A.B.A.; and the Ragon Institute Summer Program Fellowship to X.W.
Author information
Authors and Affiliations
Contributions
S.M.B. and D.S.K. conceived the overall study and guided it throughout, with input from B.M.W., E.P.B. and C.M.M.; S.M.B., N.A.M. and J.K.R. performed primary bacterial isolations; S.M.B., N.A.M., J.F.F., B.M.W., A.J.M., X.W., N.C. and C.M.M. contributed to media design and production and/or bacterial growth and inhibition experiments; B.M.W. and E.P.B. synthesized labelled glutathione; B.M.W., S.M.B., N.A.M. and E.P.B. designed, performed and/or analysed measurements of media composition and isotopic tracing experiments; S.M.B., N.A.M. and J.X. performed nucleic acid extractions and sequencing; S.M.B. performed bacterial 16S rRNA gene sequencing analysis; M.R.H. and D.A.R. performed bacterial isolate genomic and metagenomic sequence analysis and assembly, genome catalogue development, and phylogenetic reconstructions; S.M.B., M.R.H., F.A.H. and B.M.W. conceived and/or performed genomic pathway analysis; S.M.B. and A.B.A. performed analysis of in vivo metabolite data; K.L.D., M.D., T.G., F.X.C., T.N., N.I., S.M.B., N.X., M.S.G. and D.S.K. contributed to clinical cohort design, cohort performance and/or sample acquisition and processing efforts; S.M.B., B.M.W., M.R.H., N.A.M. and D.S.K. wrote the paper, and all authors reviewed, offered input to the writing and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Lindi Masson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 L-cysteine supplementation supports L. iners growth in Lactobacillus MRS broth, augmented by L-glutamine.
a,b Growth of L. crispatus, L. iners, and G. vaginalis at 24 hours incubation in MRS broth (BD DIFCO) ± supplementation with 2% IsoVitaleX (“Iso”) or with the indicated sub-pools of IsoVitaleX components. c, Growth in MRS broth supplemented with 2% IsoVitaleX or various combinations of the nutrients in “Pool 2”. d, Growth of L. iners at 24 hours in MRS broth + L-Gln (1.1 mM) supplemented with varying concentrations of L-Cys or e, in MRS broth + L-Cys (4 mM) supplemented with varying concentrations of L-Gln. Experiments in (a-e) all used BD DIFCO-formulated MRS broth base. (f) Growth of L. iners in Hardy Criterion-formulated MRS (“HMRS”) broth supplemented with IsoVitaleX 2% v/v, L-Cys (4 mM), and/or L-Gln (1.1 mM) produced similar results, although with a substantially longer lag phase. Each plot depicts median (± range) for 3 replicates per condition and each plot is representative 1 of ≥2 independent experiments per strain and media condition except (a), which was un-replicated.
Extended Data Fig. 2 Phylogeny and per-genome gene content of genomes and MAGs within FGT Lactobacillus genome catalogs.
a, Phylogenetic tree of L. iners isolate genomes and MAGs as in Fig. 1b, plus isolate genomes of L. crispatus (n = 182), L. jensenii (n = 39), L. gasseri (n = 28), and L. vaginalis (n = 8). Isolates further experimentally studied in this work are indicated. The tree depicts only genome assemblies exceeding certain quality thresholds to ensure robustness of the phylogenetic reconstruction (see Methods); additional strains and genomes were included in other analyses. b, Number of genes per genome for each genome retrieved from RefSeq (“Reference Genome”), previously unreported isolate genome (“Novel Genome”), or MAG within the Lactobacillus genome catalogs analyzed in Figs. 2b and 4b and Extended Data Fig. 6a. All MAGs present in the analysis represent previously unreported assemblies (Hayward et al, manuscript in preparation, see additional information in Methods and Supplementary Tables 4-11). To maximize comprehensiveness of the pan-genomes, the catalogs included high- and medium-quality genomes and MAGs, defined as assemblies with minimum estimated completeness >50% (some of which are classified as partial assemblies by NCBI size criteria) and maximum estimated contamination <10%. Gene count analysis excludes gene sequences observed in only 1 genome or MAG per species to eliminate singleton contaminating sequences within individual genome assemblies. Box center lines, edges, and whiskers signify the median, interquartile range (IQR), minima and maxima respectively.
Extended Data Fig. 3 In vivo association of BV status and vaginal Cys concentrations with microbiota composition.
a, Relationship between Nugent score-based BV status and cervicotype among the 53 women depicted in Fig. 3a. BV status and cervicotype were significantly associated (P = 1.902 ×10-11; two-sided Fisher’s Exact Test). b, Two-tailed Spearman correlation between relative Cys concentrations in cervicovaginal lavage (CVL) fluid and relative abundances of the species L. iners, and L. crispatus among the 142 women depicted in Fig. 3b-f, showing correlation coefficients (ρ) with unadjusted p-values. Linear regression lines (solid blue) with 95% confidence intervals calculated based on log-transformed abundances and concentrations are shown to assist visualization (L. crispatus: y = 0.77 + 0.25x; L. iners: y = 0.30 + 0.22x). The red dotted line represents the bacterial limit of detection (L.D.). c,d Per-sample relative abundances and cohort-level prevalence (fraction of samples from the cohort in which each taxon was detected) for each genus (c) or species (d) with ≥50% prevalence (panels correspond to main Figs. 3e and f, respectively). Purple and blue lines respectively represent median and interquartile range of relative abundances for each taxon.
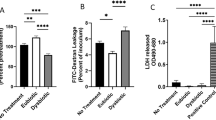
Extended Data Fig. 4 Vaginal concentrations of the Cys-containing peptides reduced glutathione (GSH) and cysteinylglycine (Cys-Gly) in cervicovaginal fluid are higher in women without BV and correlate with Lactobacillus dominance of the microbiota.
a,b Relative concentrations of (a) GSH and (b) Cys-Gly by BV status in CVL fluid from the 53 women in (Fig. 3a). c,d Relative concentrations of (c) GSH and (d) Cys-Gly by cervicotype in CVL fluid from the 142 women depicted in (Fig. 3b,c). In (a-d) the red dotted line represents the metabolite limit of detection (L.D.). For samples in which an analyte was below the L.D., concentrations were imputed at 0.5 x L.D. Log-transformed concentrations were not normally distributed due to the imputed values, so between-group differences were determined via Kruskal-Wallis test with post-hoc Dunn’s test, adjusting for multiple comparisons using the Bonferroni method. All significant pairwise differences are displayed. a No BV-BV: P = 5.7 ×10−7; No BV-Intermediate: P = 0.0037. b No BV-BV: P = 0,0032. c CT1-CT4: P = 7.7 ×10−5; CT2-CT3: P = 0.0292; CT2-CT4: P = 2.9 ×10−11. d CT1-CT4: P = 0.0191; CT2-CT4: P = 0.00014. Box center lines, edges, and whiskers signify the median, IQR, minima and maxima respectively. e,f Forest plots depicting Spearman correlation coefficients (ρ) between concentrations of GSH and Cys-Gly and relative abundances of each bacterial genus (e) or species (f) detected at >50% prevalence in the cohort (n = 142). P-values and confidence intervals in e,f were adjusted for multiple comparisons using the Bonferroni method at significance level 0.05/n (full statistical results in Supplementary Tables 14 & 15). Significance is depicted for adjusted p-values as * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.
Extended Data Fig. 5 Cys and Cys-containing molecules in MRS exist primarily as mixed disulfides and addition of chemical reducing agents permits L. iners growth.
a, Growth of L. iners and L. crispatus at 28 hours in MRSQ broth supplemented as indicated with the reduced thiols L-Cys, D-Cys (the non-physiological enantiomer of L-Cys), GSH, or homocysteine (each 4 mM), with their oxidized counterparts L-cystine, D-cystine, oxidized glutathione (GSSG), or homocystine (each 2 mM), or with the non-sulfur-containing reducing agent Tris(2-carboxyethyl)phosphine (TCEP; 4 mM), H2O2 (0.4 mM), or L-cystine + H2O2. b, Concentrations of reduced Cys (baseline median concentration 1.11 μM) and glutathione (GSH; baseline median concentration 1.70 μM) in MRSQ broth supplemented with the oxidizing agent H2O2 (0.4 mM), the reducing agents TCEP or homocysteine (each 4 mM), or homocysteine’s oxidized counterpart homocystine (2 mM). c, Growth at 7 days of L. crispatus and L. iners in HMRS broth with 1.1 mM L-Gln (“HMRSQ”) supplemented as indicated with L-Cys, L-cystine, TCEP, homocysteine, or homocystine at the above concentrations. All plots depict median ± range for 3 replicates per condition and each plot is representative of 1 of ≥2 independent experiments per strain and media condition. Bar coloring highlights the pairing of media conditions with each thiol-containing reducing agent and its oxidized counterpart.
Extended Data Fig. 6 FGT Lactobacillus genomes lack predicted alternate Cys and GSH transporters and L. iners is selectively inhibited by cystine uptake inhibitors.
a, Predicted presence of the branched-chain amino acid transport locus livFGHKM (which has low-affinity Cys transport activity in E. coli) and the glutathione transport locus gsiABCD in isolate genomes and MAGs of common FGT Lactobacillus species (n = number of genomes. Detailed statistics are in Supplementary Table 12). b, Selective growth inhibition of L. iners in MRSQ broth with or without L-Cys (4 mM) and varying concentrations of the cystine uptake inhibitor seleno-DL-cystine (SDLC). c, Growth of L. crispatus and L. iners in HMRSQ broth with or without L-Cys (4 mM) ± SMC. d, Growth inhibition of L. crispatus and L. iners by SMC (128 mM) or SDLC (2 mM) in MRSQ supplemented with L-cystine (2 mM) or D-Cys, TCEP, or GSH (4 mM each). For each growth additive, percentage growth in in presence of inhibitor was calculated relative to median growth in broth containing that additive without inhibitor. Plots in b-d depict median ± range for 3 replicates per condition and each plot is representative 1 of ≥2 independent experiments per strain and media condition.
Extended Data Fig. 7 Sample preparation and controls for L. iners / L. crispatus growth competition assays.
a, Mono-culture growth of L. crispatus (strain FRESH1) and 3 representative L. iners strains at 28 hrs incubation in MRSQ broth with or without L-Cys (4 mM) and varying concentrations of the cystine uptake inhibitor S-methyl-L-cysteine (SMC), exhibiting expected growth patterns for the respective species. Plots depict median ± range for 3 replicates per condition and each plot is representative 1 of ≥2 independent experiments per strain and media condition. The competition assays between L. iners and L. crispatus depicted in Fig. 5a were prepared by mixing the input inocula from each of these L. iners monocultures pairwise with the input inoculum for the L. crispatus monoculture at the colony-forming unit (c.f.u.) ratios listed in the legend of Fig. 5a. b, Bacterial 16 S rRNA gene read counts in cultured samples (n = 105 individual cultures), with negligible background read counts in blank growth media controls (n = 7 controls, one each per media type) and extraction controls (n = 7 controls) associated with the competition experiments in main Fig. 5a. (Control sample reactions were included in the sequencing library despite absence of visible PCR bands on agarose gel electrophoresis.) Box center lines, edges, and whiskers signify the median, IQR, minima and maxima respectively.
Extended Data Fig. 8 Development of “S-broth” and controls for mock BV-like community growth experiments.
a, Monoculture growth at 48 hours of experimental US (strain “233”) and South African (strains FRESH1 and FRESH2) L. crispatus strains, as well as L. iners, G. vaginalis, Prevotella bivia, Prevotella disiens, and Sneathia sanguinegens strains in various broth media including MRSQ broth + L-Cys (4 mM), NYCIII broth with or without 2% IsoVitaleX plus 5% Vitamin K1-Hemin solution (“IHK”) and/or Tween-80 (1 g/L), and in “S-broth” (see Methods) with or without Tween-80 (1 g/L). S-broth + Tween was used in subsequent experiments. (Detailed strain information is in Supplementary Table 3). Plots depict median ± range for 3 replicates per condition and each plot is representative 1 of ≥2 independent experiments per strain and media condition. b, Bacterial 16 S rRNA gene read counts in mock mixed communities (n = 72 individual cultures) and pure strain monocultures (n = 7), with negligible background read counts in blank growth media controls (n = 4 controls, one each per media type) and extraction controls (n = 2 controls) associated with the experiments in Fig. 5c,d. (Control sample reactions were included in the sequencing library despite absence of visible PCR bands on agarose gel electrophoresis.) Box center lines, edges, and whiskers signify the median, IQR, minima and maxima respectively. c, Confirmation of identity of input strains in mock BV-like communities by 16 S rRNA gene sequencing of bacterial monoculture controls, prepared from the same input inocula used for the bacterial mock communities shown in Fig. 5c,d.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and legends for Tables 1–20.
Supplementary Table
Supplementary Tables 1–20.
Source data
Source Data Fig. 1
Numerical and statistical source data for Fig. 1a,c and Newick-format phylogenetic tree structure and tree metadata for Fig. 1b.
Source Data Fig. 2
Numerical and statistical source data for Fig. 2.
Source Data Fig. 3
Numerical and statistical source data for Fig. 3.
Source Data Fig. 4
Numerical and statistical source data for Fig. 4.
Source Data Fig. 5
Numerical and statistical source data for Fig. 5.
Source Data Extended Data Fig. 1
Numerical and statistical source data for Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Newick-format phylogenetic tree structure and tree metadata for Extended Data Fig. 2a, and numerical and statistical source data for Extended Data Fig. 2b.
Source Data Extended Data Fig. 3
Numerical and statistical source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Numerical and statistical source data for Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Numerical and statistical source data for Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Numerical and statistical source data for Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Numerical and statistical source data for Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Numerical and statistical source data for Extended Data Fig. 8.
Rights and permissions
About this article
Cite this article
Bloom, S.M., Mafunda, N.A., Woolston, B.M. et al. Cysteine dependence of Lactobacillus iners is a potential therapeutic target for vaginal microbiota modulation. Nat Microbiol 7, 434–450 (2022). https://doi.org/10.1038/s41564-022-01070-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01070-7
This article is cited by
-
Culturomics reveals a hidden world of vaginal microbiota with the isolation of 206 bacteria from a single vaginal sample
Archives of Microbiology (2024)
-
Atlas of mRNA translation and decay for bacteria
Nature Microbiology (2023)
-
The association between homocysteine and bacterial vaginosis: results from NHANES 2001–2004
Scientific Reports (2023)
-
Systematic mining of the human microbiome identifies antimicrobial peptides with diverse activity spectra
Nature Microbiology (2023)
-
Trait biases in microbial reference genomes
Scientific Data (2023)