Abstract
The major transmission route for SARS-CoV-2 is airborne. However, previous studies could not elucidate the contribution between large droplets and aerosol transmission of SARS-CoV-2 and its variants. Here, we designed and validated an optimized transmission caging setup, which allows for the assessment of aerosol transmission efficiency at various distances. At a distance of 2 m, only particles of <5 μm traversed between cages. Using this setup, we investigated the relative efficiency of aerosol transmission between the SARS-CoV-2 Alpha variant (B.1.1.7) and lineage A in Syrian hamsters. Aerosol transmission of both variants was confirmed in all sentinels after 24 h of exposure as demonstrated by respiratory virus shedding and seroconversion. Productive transmission also occurred after 1 h of exposure, highlighting the efficiency of this transmission route. Interestingly, after donors were infected with a mix of both variants, the Alpha variant outcompeted the lineage A variant in an airborne transmission chain. Overall, these data indicate that a lower infectious dose of the Alpha variant, compared to lineage A, could be sufficient for successful transmission. This highlights the continuous need to assess emerging variants and the development for pre-emptive transmission mitigation strategies.
Similar content being viewed by others
Main
Epidemiological data suggest that the principal mode of infection with SARS-CoV-2 is via airborne and large droplet transmission1,2,3,4,5. Respiratory aerosols range from 0.1 to 100 µm (aerodynamic diameter), with the fraction of particles <5 µm being referred to as ‘fine aerosols’6. Fine aerosols remain suspended for longer periods and can penetrate all the way down into the alveolar space of the human respiratory tract7. For influenza A virus, studies have elucidated the airborne potential and relative contribution of large droplets versus aerosols and the site of viral exposure and shedding8,9,10,11. Data for SARS-CoV-2 is currently unavailable.
Variants of concern (VOCs) are defined by phenotypic changes including enhanced transmission12,13. Transmissibility is a function of infectiousness, susceptibility, contact patterns between individuals and environmental stress on the pathogen during transmission14. No study so far has demonstrated the potential of SARS-CoV-2 for fine aerosol transmission. In this study, we specifically designed cages to model aerosol transmission over a 2-m distance, at which only particles <5 µm traverse. We showed highly efficient aerosol transmission of SARS-CoV-2 at 2 m within 1 h of exposure. Lastly, we demonstrated increased airborne transmission competitiveness of B.1.1.7 over a lineage A variant.
Design and validation of hamster aerosol transmission cages
Demonstration of true aerosol transmission of SARS-CoV-2 should only include particles <5 µm over short and longer distances and in the absence of any other potential transmission routes such as fomite or direct contact. We designed and validated a caging system to study the relationship between particle size and distance. The design consisted of two rodent cages connected via a connection tube that allowed airflow but no direct animal contact. The distance between donor and sentinel cage could be varied (16.5, 106 or 200 cm) (Supplementary Fig. 1a,b). The air velocity generated by the airflow through the connection tube averaged at 327, 370 and 420 cm min−1 for the 16.5, 106 and 200 cm distances, respectively (Supplementary Table 1). This allowed for 30 cage air changes per hour.
We next validated the caging design using an aerodynamic particle sizer spectrometer (TSI) to analyse the aerodynamic size of particles (dynamic range <0.5–20 µm) traversing from donor to sentinel cage15. The aerodynamic particle size of a particle is the particle size regardless of shape and material density that has the same settling rate as a spherical droplet of water. It is the principal parameter used for characterizing respiratory deposition7. Therefore, a 5 µm aerodynamical particle will act similarly to a 5 µm spherical droplet of water whether it is a from a 20% glycerol solution or a mucus-filled respiratory droplet. A 20% glycerol solution has a density of 1.05 g cm−3; therefore, the geometrical diameter of the glycerol droplets is determined by multiplying the aerodynamic particle diameter by a correction factor of 0.98 (ref. 16). Droplets and aerosols were generated in the donor cage and the particle size profile was determined at the beginning and end of the connecting tube to study the size exclusion of the respective cage set-ups. The reduction of particles was size- and distance-dependent. At a distance of 16.5 cm, relatively limited size exclusion of the generated particles was observed; ≥6.9% of particles 5–10 µm and ≥42.8% of particles ≥10 µm did not travel into the sentinel cage (Fig. 1a,d). At 106 cm between cages, an increased reduction of number of particles and size exclusion was observed; ≥70% of particles ≥5 µm did not traverse into the sentinel cage and no particles ≥10 µm were detected. Hence, while in the donor cage 4.86% of detected particles were >5 µm, in comparison the particle profile in the sentinel cage contained only 2% particles >5 µm (Fig. 1b,e). At 200 cm, we observed an almost complete size exclusion of particles ≥5 µm; ≥95% of particles 5–10 µm did not traverse and no particles ≥10 µm were detected in the sentinel cage. The composition profile of particles in the sentinel cage comprised only 0.5% particles ≥5 µm (Fig. 1c,f). These results demonstrate that we have developed a caging system to effectively investigate the impact of distance and particle size exclusion on the transmission of SARS-CoV-2. The overall absence of particles ≥10 µm and extensive reduction of particles 5–10 µm indicate that the caging system with a distance of 200 cm is suitable to study true aerosol transmission, whereas the 16.5 and 106 cm set-ups are suitable to study airborne transmission occurring via droplet, aerosols or a combination thereof.
Transmission cages were designed to model airborne transmission between Syrian hamsters at 16.5, 106 and 200 cm distances. Droplets were generated by spraying a 20% glycerol/water solution into the donor cage. The size of particles travelling between donor and sentinel cages was determined. a–c, Particle reduction by aerodynamic diameter between donor and sentinel cages at 16.5 (a), 106 (b) and 200 cm distances (c). The dotted line represents 95% reduction in particles. The aerodynamic diameter was 1–10 μm. d–f, Schematic visualization of the transmission cages at 16.5 (d), 106 (e) and 200 cm distances (f) and corresponding particle distribution detected in each donor and sentinel cage.
SARS-CoV-2 aerosol transmission over 2 m
Using the validated caging system, we first investigated short-distance airborne transmission. For each distance, 4 donor animals were inoculated intranasal with 8 × 104 median tissue culture infectious dose (TCID50) SARS-CoV-2 lineage A. After 12 h, the infected animals were placed into the donor (upstream) side of the cages and 4 sentinels were placed into the downstream cages (2:2 ratio) and were exposed for 72 h.
At 16.5 cm, SARS-CoV-2 was successfully transmitted to all sentinels at 12 h post-exposure (Fig. 2a–c). Genomic and subgenomic RNA, a marker for replicating virus, and infectious virus were found in the oropharyngeal swabs of all sentinels at 72 h post-exposure. At 106 cm, SARS-CoV-2 genomic and subgenomic RNA were detected in the oropharyngeal swabs of 1 sentinel as early as 12 h post-exposure. At 48 h post-exposure, all sentinels were positive for genomic and subgenomic RNA in oropharyngeal swabs and at 72 h post-exposure for infectious virus (Fig. 2d–f). At 200 cm, no respiratory shedding was detectable in any sentinel 12 h post-exposure, but at 48 h post-exposure all sentinels were positive for genomic RNA, subgenomic RNA and infectious virus in oropharyngeal swabs (Fig. 2g–i). Additionally, at 14 d post-exposure, all sentinels from all three groups had seroconverted, as demonstrated by high antibody titres against SARS-CoV-2, measured by anti-spike enzyme-linked immunosorbent assay (ELISA) (Supplementary Table 2). These data demonstrate the ability of SARS-CoV-2 to transmit over long and short distances.
Donor Syrian hamsters were inoculated with 8 × 104 TCID50 SARS-CoV-2. After 12 h, donors were introduced to the upstream cage and sentinels (2:2 ratio) into the downstream cage. Exposure was continued for 3 d. To demonstrate transmission, sentinels were monitored for start and continuation of respiratory shedding. a–c, Viral load in the oropharyngeal swabs of sentinels was measured by genomic RNA (a), subgenomic RNA (b) and infectious virus (c) collected at 12, 24 and 48 h post-exposure to the donors. Exposure at 16.5 cm distance. d–f, Exposure at 106 cm distance. Viral load in the oropharyngeal swabs of sentinels was measured by genomic RNA (d), subgenomic RNA (e) and infectious virus (f) collected at 12, 24 and 48 h post-exposure to the donors. g–i, Exposure at 200 cm distance. Viral load in the oropharyngeal swabs of sentinels was measured by genomic RNA (g), subgenomic RNA (h) and infectious virus (i) collected at 12, 24 and 48 h post-exposure to the donors. The truncated violin plots depict the median, quantiles and individuals, n = 4.
Increased cellular entry and respiratory shedding of B.1.1.7 variant in hamsters
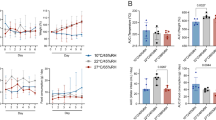
We then compared the airborne transmission kinetics of the B.1.1.7 variant with the prototype lineage A virus. First, we assessed the suitability of the Syrian hamster to compare SARS-CoV-2 variant transmission17. Differences in the amino acid sequence were visualized by mapping onto a structure of the SARS-CoV-2 spike protein in complex with human angiotensin-converting enzyme 2 (ACE2; Protein Data Bank (PDB) ID: 6M0J). At position 501 of the B.1.1.7 spike receptor-binding domain (RBD), the asparagine residue is substituted by tyrosine. This substitution has been proposed to increase binding affinity to (human) ACE2 via increased interactions with residues on ACE2 by stacking of aromatic side chains and hydrogen bond interactions16. A sequence alignment between human and hamster ACE2 reveals that two residues differ in the interface with SARS-CoV-2 RBD. At positions 34 and 82, histidine and methionine are replaced by glutamine and asparagine, respectively in the hamster ACE2 (Fig. 3a,b). It is possible that subtle changes resulting from these substitutions in hamster ACE2, not captured in this visualization, could affect the binding interactions with the B.1.1.7 variant. Interestingly, the substitution to asparagine at position 82 introduces an N-linked glycosylation sequon (NYS). This residue is located at the periphery of the interface and makes minor contributions in the human complex. To evaluate whether these sequence differences in hamster ACE2 may affect viral entry, we also directly compared viral entry using a vesicular stomatitis virus (VSV) pseudotype entry assay. No significant difference in entry between human and hamster ACE2 with either lineage A or B.1.1.7 was observed. For both human and hamster ACE2, B.1.1.7 demonstrated significantly increased entry compared to the lineage A variant (human ACE2 median lineage A B.1.1.7 = 156.8 out of 256 (relative entry to no-spike), P < 0.0001; hamster ACE2 median lineage A B.1.1.7 = 144.6 out of 197.5 (relative entry to no-spike), P = 0.003, n = 14, Mann–Whitney U-test) (Fig. 3c).
a, Differences between hamster and human ACE2 and between lineage A and B.1.1.7 SARS-CoV-2 RBD are shown on the structure of the human ACE2–RBD complex (PDB ID: 6M0J53). Representation of human ACE2 is coloured black and RBD is coloured grey. Side chains of the differing residues and surrounding residues involved in the interface are shown as sticks. The boxes show close-up views highlighting residues that differ between the two RBDs and between human and hamster ACE2 within the interface. The side chains of residues at the N501Y substitution in the B.1.1.7 variant RBD, as well as the hamster ACE2 H34Q and M82N substitutions, are coloured red and shown superposed to the side chain of the original residue. b, Amino acid sequence alignments of human ACE2 (BAB40370.1), hamster ACE2 (XP_005074266.1) and SARS-CoV-2 RBD from the A lineage strain and B.1.1.7 variant (bottom). Residues involved in the RBD–ACE2 interaction are coloured orange. Residues that participate in intermolecular hydrogen bonding or salt bridges are shown as black dots. ACE2 residues that differ between hamster and human within the interface are outlined with a box and highlighted in (a). RBD residue 501 has been highlighted with a red box. c, The relative entry of lineage A and B.1.1.7 variant to no-spike control for human and hamster ACE2 is shown. The truncated violin plots depict the median, quantiles and individuals, n = 14, Mann–Whitney U-test, two-tailed, P = <0.0001 (human ACE2) and P = 0.0030 (hamster ACE2). d, Relative weight loss in hamsters after lineage A or B.1.1.7 variant inoculation. The graph shows the median and 95% CI, n = 10. e, Viral load as measured by genomic and subgenomic RNA in lungs collected at day 5 post-inoculation. The truncated violin plots depict the median, quantiles and individuals, n = 5, ordinary two-way ANOVA followed by Sidak’s multiple comparisons test, P = 0.0045 (subgenomic). f, Infectious virus determined by titration in the lungs. The truncated violin plots depict the median, quantiles and individuals, n = 5. g, Binding antibodies against the spike protein of SARS-CoV-2 in serum obtained 14 d post-inoculation. The truncated violin plots depict the median, quantiles and individuals, n = 5, two-tailed Mann–Whitney U-test, P = 0.0397. ELISA was performed once. h, Viral load as measured by subgenomic RNA in oropharyngeal swabs. The truncated violin plots depict the median, quantiles and individuals, n = 10. i, AUC analysis of cumulative respiratory shedding. The truncated violin plots depict the median, quantiles and individuals, n = 5, two-tailed Mann–Whitney U-test, P = 0.0079. Blue, lineage A; red, B.1.1.7, n=5. P values are indicated were appropriate.
We next investigated if infection of hamsters with the B.1.1.7 variant translated to differences in viral replication and shedding dynamics in vivo.
Hamsters (n = 10) were inoculated intranasally with 102 TCID50 of SARS-CoV-2 lineage A or B.1.1.7. variant. Regardless of variant, weight loss was observed in all animals with a maximum at 7 d post-inoculation, after which animals began to recover (Fig. 3d, n = 5, median weight loss lineage A/B.1.1.7 = 7.5/8.2%). Five out of ten hamsters per group were euthanized at 5 d post-inoculation and lung tissue was collected to assess viral replication in the lower respiratory tract. The lung tissue of animals inoculated with B.1.1.7 contained higher levels of genomic RNA and significantly higher levels of subgenomic RNA (Fig. 3e, n = 5, two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons test, median lineage A/B.1.1.7 = 9.9/10.5 log10 copies g−1 and P = 0.0614; median lineage A/B.1.1.7 = 9.2/10 log10 copies g−1 and P = 0.0045, respectively). Infectious virus titres in the lungs were not significantly different between variants (Fig. 3f, n = 5 median lineage A/B.1.1.7 = 6.3/6.5 log10 TCID50 g−1). At 14 d post-inoculation, all remaining animals had seroconverted. Anti-spike immunoglobulin G (IgG) ELISA titres were significantly increased in animals inoculated with B.1.1.7 (Fig. 3g, n = 5, Mann–Whitney U-test, median lineage A/B.1.1.7 = 102,400/204,800 and P = 0.0394). Next, we studied differences in shedding from the upper respiratory tract. Subgenomic RNA was detected in 2 animals at 12 h post-inoculation with the lineage A variant and in 1 animal inoculated with the B.1.1.7. variant. Subgenomic RNA was detected at similar levels for both groups at 24 h post-inoculation (Fig. 3h). At 3 d post-inoculation, a significant increase in subgenomic RNA was seen in animals inoculated with B.1.1.7. In both groups, subgenomic RNA levels from oropharyngeal swabs started to drop at 5 d post-inoculation; levels in the B.1.1.7 animals remained somewhat higher (3 d post-inoculation median lineage A/B.1.1.7 = 6.9/7.6 and 5 d post-inoculation median lineage A/B.1.1.7 = 6.0/6.5 copies ml−1 (log10)). This translated to a significant difference when comparing the cumulative shedding until 5 d post-inoculation (Fig. 3i, area under the curve (AUC), n = 5, Mann–Whitney U-test, median lineage A/B.1.1.7 = 726/770 cumulative copies ml−1 (log10) and P = 0.0079).
Efficient aerosol transmission with B.1.1.7
We repeated the aerosol transmission experiment at 106 and 200 cm as described above for B.1.1.7. Aerosol transmission of B.1.1.7 was equally as efficient as for lineage A; all sentinels demonstrated respiratory shedding and seroconversion (Fig. 4a–f and Supplementary Table 1).
Comparison of the aerosol transmission efficiency of the lineage A and B.1.1.7 SARS-CoV-2 variants in the Syrian hamster. a–c, Exposure at 106 cm distance. Donor Syrian hamsters were inoculated with 8 × 104 TCID50 SARS-CoV-2 B.1.1.7 variant. After 12 h, donors were introduced to the upstream cage and sentinels (2:2 ratio) into the downstream cage. Exposure was continued for 3 d. To demonstrate transmission, sentinels were monitored for the start and continuation of respiratory shedding. The viral load in the oropharyngeal swabs of sentinels was measured by genomic RNA (a), subgenomic RNA (b) and infectious virus (c) collected at 12, 24 and 48 h post-exposure to the donors. d–f, Exposure at 200 cm distance. The viral load in the oropharyngeal swabs of sentinels was measured by genomic RNA (d), subgenomic RNA (e) and infectious virus (f) collected at 12, 24 and 48 h post-exposure to the donors. g–i, Donor Syrian hamsters were inoculated with 8 × 104 TCID50 SARS-CoV-2 B.1.1.7 variant or lineage A (n = 4, respectively). After 12 h, donors were introduced to the upstream cage and sentinels (2:2 ratio) into the downstream cage. Exposure was limited to 4 h. Genomic RNA (g), subgenomic RNA (h) and infectious virus (i) in oropharyngeal swabs were collected at 24, 48 and 72 h post-exposure to the donors. j–l, Exposure was limited to 1 h for B.1.1.7 and lineage A (n = 4, respectively). Genomic RNA (j), subgenomic RNA (k) and infectious virus (l) in the oropharyngeal swabs were collected at 24, 48 and 72 h post-exposure to the donors. m–o, The viral load in the oropharyngeal swabs of donors was measured by genomic RNA (m), subgenomic RNA (n) and infectious virus (o) collected 24 h post-inoculation. The truncated violin plots depict the median, quantiles and individuals; blue, lineage A, red, B.1.1.7, n = 4 for each variant, two-way ANOVA followed by Sidak’s multiple comparisons test. P values are indicated were appropriate.
We next set out to determine the transmission efficiency within a limited exposure window of either 1 or 4 h at 200 cm distance. First, four donor animals were inoculated intranasally with 8 × 104 TCID50 SARS-CoV-2 lineage A and four donors with B.1.1.7. Sentinels were exposed in a 2:2 ratio at 12 h post-inoculation for 4 h. Transmission via aerosols occurred even when time of exposure was limited for both variants. Both genomic and subgenomic RNA shedding were detected 24 h after exposure in oropharyngeal swabs of 3 out of 4 sentinel animals exposed to B.1.1.7 and lineage A (Fig. 4g,h). At 3 d post-exposure, all sentinels displayed genomic and subgenomic RNA shedding; infectious virus was detected in all sentinels exposed to lineage A and 3 out of 4 exposed to B.1.1.7 (Fig. 4i).
Exposure time was limited to 1 h. Genomic RNA was detected 24 h post-exposure in oropharyngeal swabs of 3 out of 4 sentinels exposed to B.1.1.7 and 2 out of 4 sentinels exposed to lineage A (Fig. 4j–l). Subgenomic RNA was detected in the oropharyngeal swabs of two out of four sentinels exposed to B.1.1.7 and two out of four sentinels for lineage A. At 3 d post-exposure, while all sentinels exposed to B.1.1.7 were positive for genomic RNA, subgenomic RNA and infectious virus, viral RNA was only detected in 2 of 4 sentinels exposed to lineage A. Viral loads in swabs did not differ significantly between the two variants. To ensure the differences observed in transmission were not due to increased donor shedding, we compared viral loads in oropharyngeal swabs taken from donor animals after exposure. B.1.1.7 did not significantly impact the respiratory shedding of the donors at this time point (Fig. 4m–o, n = 4, two-way ANOVA followed by Sidak’s multiple comparisons test; 4 h: genomic RNA P = 0.8737, subgenomic RNA P = 0.1049 and infectious virus P = 0.3038; 1 h: genomic RNA P = 0.6853, subgenomic RNA P = 0.2450 and infectious virus P = 0.0793). The same experiment was repeated on day 3 after inoculation of donors. No transmission occurred at this time point; we did not observe genomic RNA in oropharyngeal swabs of any sentinel on consecutive days and no subgenomic RNA was detected in any swab taken during 3 d post-exposure (Supplementary Fig. 2). These data suggest that aerosol transmission for B.1.1.7 may be more efficient compared to lineage A and may be independent of the amount of virus shed by the donor.
B.1.1.7 variant demonstrates increased airborne transmission competitiveness
We then employed the 16.5 cm cage system to conduct a transmission chain study. Donor animals (n = 8) were inoculated intranasally with 1 × 102 TCID50 SARS-CoV-2 (1:1 lineage A:B.1.1.7 mixture). Dual infection presented with comparable weight loss and shedding profile to inoculation with either variant (Supplementary Fig. 3a,b). After 12 h, donors were cohoused with 8 sentinels (sentinels 1) (2:2 ratio) for 24 h (Fig. 5a). Immediately after, the 8 sentinels were cohoused with 8 new sentinels (sentinels 2) (2:2 ratio) for 24 h and donor animals were relocated to normal caging. This sequence was repeated for sentinels 3. For each round, the previous sentinels were housed in the upstream cage and were the new donors. We assessed transmission by measuring viral RNA in oropharyngeal swabs taken from all animals at 2 d post-infection/post-exposure. While all donor animals (median genomic RNA = 7.3 copies ml−1 (log10), median subgenomic RNA = 7.0 copies ml−1 (log10)) and all sentinels 1 (median genomic RNA = 7.0 copies ml−1 (log10), median subgenomic RNA = 6.8 copies ml−1 (log10)) demonstrated robust shedding, viral RNA could only be detected in 4 out of 8 sentinels 2 (median genomic RNA = 2.5 copies ml−1 (log10), median subgenomic RNA = 1.8 copies ml−1 (log10)) and in 1 sentinels 3 animal (Fig. 5b,c). We compared infectious virus titres in the swabs. While all donor animals (median = 4.25 TCID50 (log10)) and all sentinels 1 had high infectious virus titres (median = 4.5 TCID50 (log10)), infectious virus could only be detected in 4 sentinels 2 (median = 0.9 TCID50 (log10)) and no sentinels 3 (Fig. 5d). We then compared the viral loads in the lungs of these animals at 5 d post-exposure. While all donor animals (median subgenomic RNA = 10.0 copies g−1 (log10)) and all sentinels 1 had high subgenomic RNA levels in the lung (median subgenomic RNA = 9.7 copies ml−1 (log10)), viral RNA could be detected in sentinels 2 at a lower level (median subgenomic RNA = 7.1 copies ml−1 (log10)) and no subgenomic RNA was detected in any sentinels 3 (Fig. 5e). Grossly, lungs from sentinels 1 demonstrated SARS-CoV-2 infection associated pathology as described previously18,19,20. Pathological lesions were only seen in three sentinels 2 and no sentinels 3 (Fig. 4f, Supplementary Table 3 and Supplementary Fig. 3c). This suggests that transmission very early after exposure may be restricted and that not all animals were able to efficiently transmit the virus to the next round of naïve sentinels.
Donor animals (n = 8) were inoculated with both lineage A and B.1.1.7 variant with 102 TCID50 via the intranasal route (1:1 ratio) and three groups of sentinels (sentinels 1, 2 and 3) were exposed subsequently at a 16.5 cm distance. a, Schematic visualization of the transmission chain design. Animals were exposed at a 2:2 ratio; exposure occurred on consecutive days and lasted for 24 h for each chain link. b,c, Respiratory shedding measured by viral load in oropharyngeal swabs, measured by genomic (b) and subgenomic RNA (c) on day 2 post-exposure. The truncated violin plots depict the median, quantiles and individuals, n = 8. d, Corresponding infectious virus in oropharyngeal swabs, measured by titration. The truncated violin plots depict the median, quantiles and individuals, n = 8. e, Corresponding infectious virus in lungs sampled 5 d post-exposure, measured by titration. The truncated violin plots depict the median, quantiles and individuals, n = 8. f, Gross pathology of lungs at day 5 post-exposure. g, Percentage of B.1.1.7 detected in oropharyngeal swabs taken at day 2 post-exposure for each individual donor and sentinel, determined by deep sequencing. The pie charts depict individual animals. Red, B.1.1.7; blue, lineage A; grey, no viral RNA present in sample; transparent colour, duplex RT–qPCR confirmed viral RNA presence in the sample but sequencing was unsuccessful due to low RNA quality. h, Percentage of B.1.1.7 detected in lungs taken at day 5 post-exposure for each individual donor and sentinel, determined by deep sequencing. The pie charts depict individual animals. Red, B.1.1.7; blue, lineage A; grey, no viral RNA present in the sample.
To determine the competitiveness of the variants, we analysed the relative composition of the two viruses in the swabs using next-generation sequencing (NGS) and compared the percentage of B.1.1.7/lineage A at 2 d post-exposure in oral swab samples and at 5 d post-exposure in lungs (Fig. 5g,h). We observed 1 donor with increased amounts of lineage A variant (55%), while in the remaining 7 animals the B.1.1.7 variant was increased (range 54–74%). After the first airborne transmission sequence, 2 sentinels shed increased amounts of lineage A variant (55 and 84%), while the remaining 6 shed more B.1.1.7, five of which shed nearly exclusively B.1.1.7 (>96%). After the second round of the airborne transmission sequence, three out of four sentinel animals shed exclusively B.1.1.7 and one animal shed exclusively lineage A. Due to low amounts of viral RNA, two sentinels in the sentinels 2 and one sentinel in the sentinels 3 groups could not be successfully sequenced. We analysed these samples by duplex quantitative PCR with reverse transcription (RT–qPCR) applying a modified \(2^{-\Delta\Delta_{\rm{CT}}}\) method (Supplementary Table 4). One animal in the sentinels 2 group only shed B.1.1.7 (no lineage A PCR positivity). The other shed nearly exclusively lineage A (0.0007-fold increase of B.1.1.7), while, interestingly, the transmission event of this animal to the sentinels 3 animal was exclusively B.1.1.7. We sequenced viral RNA extracted from lung samples collected at 5 d post-exposure for all donors, sentinels 1 and sentinels 2. One donor had increased amounts of lineage A variant (82%), while in the remaining 7 animals the B.1.1.7 variant was increased in the lungs (range 73–85%). In the sentinels 1 group, increased amounts of lineage A variant were found in 2 animals (72 and 99%), while B.1.1.7 was increased in the others (65–100%). In the sentinels 2 group, viral RNA sequences in two animals were 100% lineage A and in the remaining 4 animals exclusively B.1.1.7 (99–100%). Taken together, B.1.1.7 demonstrated increased competitiveness. After 10 out of 13 airborne transmission events, B.1.1.7 outcompeted lineage A in the upper respiratory tract (swab samples); only in 3 events infection with lineage A was established as the dominant variant. In the lower respiratory tract, B.1.1.7 outcompeted lineage A in 10 out of 14 events.
Discussion
Epidemiological studies in humans and experimental studies in ferrets strongly suggest that aerosol transmission occurs and plays a major role in driving the SARS-CoV-2 pandemic21,22,23,24,25. Yet conclusive experimental proof of aerosol transmission with a particle diameter <5 µm26 in the absence of other transmission routes has not been provided27. In this study, we demonstrated efficient transmission of SARS-CoV-2 between Syrian hamsters via particles <5 µm over a 200 cm distance. Additionally, we present qualitative analyses of the efficiency of transmission, showing that even within 1 h transmission can occur at a distance of 200 cm between Syrian hamsters.
Whereas several SARS-CoV-2 airborne transmission studies in hamsters and ferrets have been performed, none of these studies were able to differentiate between large and small droplet transmission18,24,28,29,30. Within the currently described transmission caging, only 2% and 0.5% of particles found in the sentinel side were ≥5 µm at the 106 and 200 cm distances, respectively, strongly suggesting that the transmission observed in these cages occurs by true aerosols. This is an important finding since droplets <5 µm are expected to reach the respiratory bronchioles and alveoli. While respirable aerosol (<2.5 µm), thoracic aerosol (<10 µm) and inhalable aerosol in general31 may be relevant to infection with SARS-CoV-2 (ref. 32), it has been suggested that direct deposition into the lower respiratory tract may decrease the necessary infectious dose in humans33. Indeed, previously we demonstrated that aerosol inoculation in the Syrian hamster is highly efficient (25 TCID50, particles <5 µm34) and is linked to increased disease severity due to direct deposition into the lower respiratory tract18.
Experimentally, animals were exposed to a unidirectional airflow at time points chosen for optimal donor shedding, which likely contributed to the high efficiency of aerosol transmission. However, this approximates human exposure settings, such as restaurants or office spaces.
Increased risk of airborne transmission is an important concern in the context of VOCs. VOC B.1.1.7 has been shown to exhibit increased transmission with significantly increased reproduction number and attack rates35,36. In addition, B.1.1.7. was detected in higher concentration in fine aerosols (<5 µm) but not in coarse aerosols (≥5 µm) compared to other lineages in human respiratory samples37,38. Transmission efficiency is a function of donor shedding, exposure time, sentinel susceptibility and potential environmental factors effecting stability during transmission. Our entry data shows increased entry of B.1.1.7 over lineage A hamster ACE2. Our data suggest that the increased transmission efficiency of B.1.1.7 may not be a direct result of shedding magnitude but that a lower dose of B.1.1.7 may be sufficient for transmission. Under the applied experimental restrictions (200 cm, 1 h) both lineage A and B.1.1.7 transmitted equally as efficiently. However, B.1.1.7 displayed an increased airborne transmission competitiveness in a dual infection experiment both in the lower and upper respiratory tract. In three transmission events, we could not confirm full viral sequences in oral swab samples. However, given all other events for which complete sequencing was done, it is very likely that the variant, and not a recombinant, was selected. This has also previously been shown for D614G over the lineage A variant39,40 and for B.1.1.7 over D614G39,41,42; however, these studies did not look at airborne transmission. The additional N501Y mutation is specifically predicted to increase affinity for human ACE2, partially explaining the dominance of B.1.1.7 and other new variants containing both mutations43,44. We observed that the variant profiles in the upper and lower respiratory tract were not always similar. Additional studies are needed to understand the differential tropism and determine the location of virus in the respiratory tract involved in transmission. Additional work is required to demonstrate conclusively if the increased airborne competitiveness of B.1.1.7 in the hamster model is truly a result of increased susceptibility. The increase in aerosol transmission potential of B.1.1.7 underscores the continuous need for development and implementation of non-pharmaceutical pre-emptive interventions. This is further supported by an analogous result obtained with influenza A virus, which showed than the pandemic strain of A (subtype H1N1) was much more efficient at aerosol transmission than the seasonal A (subtype H1N1) it replaced45. In the light of limited global vaccine coverage and the potential emergence of escape mutants, ventilation, air disinfection46,47, face masks and social distancing14,48 should still be considered essential tools in preventing COVID-19 exposure and mitigating transmission risk.
Methods
Ethics
All animal experiments were conducted in an American Association for Accreditation of Laboratory Animal Care internationally accredited facility and were approved by the Rocky Mountain Laboratories Institutional Care and Use Committee according to the guidelines put forth in the Guide for the Care and Use of Laboratory Animals, 8th edition, the Animal Welfare Act, U. S. Department of Agriculture and the U. S. Public Health Service Policy on the Humane Care and Use of Laboratory Animals. Work with infectious SARS-CoV-2 virus strains under biosafety level 3 conditions was approved by the Institutional Biosafety Committee (IBC). For removal of specimens from high containment areas, virus inactivation of all samples was performed according to IBC-approved standard operating procedures.
Cells and virus
SARS-CoV-2 variant B.1.1.7 (hCoV320 19/England/204820464/2020, EPI_ISL_683466) was obtained from Public Health England via BEI. SARS-CoV-2 strain nCoV-WA1-2020 (lineage A, MN985325.1) was provided by the Centers for Disease Control and Prevention. Virus propagation was performed in VeroE6 cells (University of North Carolina; provided by R. Baric) in DMEM supplemented with 2% foetal bovine serum (FBS), 1 mM L-glutamine, 50 U ml−1 penicillin and 50 μg ml−1 streptomycin (DMEM2). VeroE6 cells were maintained in DMEM supplemented with 10% FBS, 1 mM L-glutamine, 50 U ml−1 penicillin and 50 μg ml−1 streptomycin. Mycoplasma testing was performed at regular intervals. No Mycoplasma and no contaminants were detected. For sequencing from viral stocks, sequencing libraries were prepared using the Stranded Total RNA Prep Ligation with Ribo-Zero Plus Kit according to the manufacturer’s protocol (Illumina) and sequenced on an Illumina MiSeq at 2 × 150 base pair (bp) reads. No nucleotide change was found >5% for nCoV-WA1-2020. For VOC B.1.1.7, Supplementary Table 6 summarizes the mutations obtained.
Plasmids
The spike coding sequences for SARS-CoV-2 lineage A (nCoV-WA1-2020) and variant B.1.1.7 (hCoV320 19/England/204820464/2020, EPI_ISL_683466) were truncated by deleting 19 amino acids at the C terminus. The S proteins with the 19 amino acid deletion of coronaviruses were previously reported to show increased efficiency regarding incorporation into virions of VSV49,50. These sequences were codon-optimized for human cells, then appended with a 5′ kozak expression sequence (GCCACC) and 3′ tetra-glycine linker followed by nucleotides encoding a FLAG-tag sequence (DYKDDDDK). These spike sequences were synthesized and cloned into pcDNA3.1+ (GenScript). Human and hamster ACE2 (Q9BYF1.2 and GQ262794.1) were synthesized and cloned into pcDNA3.1+ (GenScript). All DNA constructs were verified by Sanger sequencing (ACGT).
Receptor transfection
BHK cells (provided by M. Bloom) were seeded in black 96-well plates and transfected the next day with 100 ng of plasmid DNA encoding human or hamster ACE2, using polyethylenimine (Polysciences). All downstream experiments were performed 24 h post-transfection.
Pseudotype production and luciferase-based cell entry assay
Pseudotype production was carried as described previously51. Briefly, plates precoated with poly-L-lysine (Sigma-Aldrich) were seeded with 293T cells (provided by S. Best) and transfected the following day with 1,200 ng of empty plasmid and 400 ng of plasmid encoding coronavirus spike or no-spike plasmid control (green fluorescent protein (GFP)). After 24 h, transfected cells were infected with VSVΔG seed particles pseudotyped with VSV-G, as described previously51,52. After 1 h of incubation with intermittent shaking at 37 °C, cells were washed 4 times and incubated in 2 ml DMEM supplemented with 2% FBS, penicillin-streptomycin and L-glutamine for 48 h. Supernatants were collected, centrifuged at 500g for 5 min, aliquoted and stored at −80 °C. BHK cells previously transfected with ACE2 plasmid of interest were inoculated with equivalent volumes of pseudotype stocks. Plates were then centrifuged at 1,200g at 4 °C for 1 h and incubated overnight at 37 °C. Approximately 18–20 h post-infection, Bright-Glo luciferase reagent (Promega Corporation) was added to each well, 1:1, and luciferase was measured. Relative entry was calculated normalizing the relative light unit (RLU) for spike pseudotypes to the plate RLU average for the no-spike control. Each figure shows the data for two technical replicates.
Structural interaction analysis
Structure modelling was performed using the human ACE2 and SARS-CoV-2 RBD crystal structure (PDB ID 6M0J)53. Mutagenesis to model the residues that differed in the B.1.1.7 RBD and hamster ACE2 was performed in COOT v.0.9.4.154. The structure figure was generated using the PyMOL Molecular Graphics System (https://www.schrodinger.com/pymol). Amino acid sequence alignments of human (BAB40370.1) and hamster ACE2 (XP_005074266.1) and of SARS-CoV-2 RBD from the linage A strain and B.1.1.7 variant were generated using Clustal Omega v.1.2.4 (http://europepmc.org/article/MED/). Residues participating in the SARS-CoV-2–ACE2 interface were noted as described by Lan et al.53.
Duplex RT–qPCR variant detection
The duplex RT–qPCR primers and probe were designed to distinguish between lineage A SARS-CoV-2 and the B.1.1.7 variant (Supplemental Table 5) in a duplex assay. The forward and reverse primers were designed to detect both variants while two probes were designed to detect either variant. Five microlitres of RNA were tested with the TaqMan Fast Virus One-Step Master Mix (Applied Biosystems) using the QuantStudio 3 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The relative fold change difference between both variants was calculated by applying the \(2^{-\Delta\Delta_{\rm{CT}}}\) method with modifications.
Inoculation experiments
Four-to-six-week-old female and male Syrian hamsters (ENVIGO) were inoculated (10 animals per virus) intranasally with either the SARS-CoV-2 strain nCoV-WA1-2020 (lineage A) or hCoV320 19/England/204820464/2020 (B.1.1.7) or a 1:1 mixture of both viruses. Intranasal inoculation was performed with 40 µl of sterile DMEM containing 1 × 102 TCID50 SARS-CoV-2. At 5 d post-inoculation, 5 hamsters from each route were euthanized and tissues were collected. The remaining 5 animals from each route were euthanized at 14 d post-inoculation for disease course assessment and shedding analysis. Hamsters were weighed daily and oropharyngeal swabs were taken on days 1, 2, 3 and 5. Swabs were collected in 1 ml DMEM with 200 U ml−1 penicillin and 200 µg ml−1 streptomycin. Hamsters were observed daily for clinical signs of disease. Necropsy and tissue sampling were performed according to IBC-approved protocols.
Aerosol cages
The aerosol transmission system consisted of two 17.78 x 27.04 x 22.86 cm (7'x 11' x 9') plastic hamster boxes (Lab Products) connected with a 7.62 cm (3') diameter tube (Supplementary Fig. 1). The boxes were modified to accept a 7.62 cm (3') plastic sanitary fitting (McMaster-Carr), which enabled the length between the boxes to be changed. The nominal tube lengths were 16.5, 106 and 200 cm. Airflow was generated with a vacuum pump (Vacuubrand) attached to the box housing the naïve animals and was controlled with a float-type metre/valve (McMaster-Carr). The airflow was adjusted for each tube length to be 30 cage changes per hour and the flow was validated before starting the experiments by timing a smoke plume through the tubes. The airflow of the original boxes was in through a filtered top and out through an exhaust port in the side of the box. To ensure proper airflow from the donor box to the naïve box, the top of the naïve box was sealed while the filter top of the donor box remained open.
To ensure the system contained aerosols, the airtightness of the system was validated with a negative pressure smoke test and a positive pressure leak test before moving into a containment laboratory. To perform the negative pressure test, the airflow was adjusted to exhaust the system at 30 cage changes per hour; smoke was generated in the donor cage with a WizardStick and escaped particulate was measured with a DustTrak DRX (TSI). To test the system under pressure, the air flow was reversed and the joints were tested using a gas leak detector.
Particle sizing
Transmission cages were modified by introducing an inlet on the side wall of the infected hamster side and sample ports on each end of the connection tube to measure particles in the air under constant airflow conditions. Particles were generated by spraying a 20% (v/v) glycerol solution with a standard spray bottle through the donor cage inlet. Particle size was measured using a Model 3321 aerodynamic particle sizer spectrometer (TSI). First, the donor cage was coated with three sprays at an interval of 30 s. The sample port was opened and a sample was analysed. Every 30 s a new spray followed and 5 samples were analysed (5 runs, each 60 s) for both donor side (primary infected side) and sentinel side. Data were collected using Aerosol Instrument Manager v.10.3 (TSI).
Aerosol transmission experiments
Transmission studies were conducted at a 2:2 ratio between donor and sentinels for each transmission scenario and virus variant with 2 separate transmission cages (n = 4 donors or 4 sentinels). Four-to-six-week-old female and male Syrian hamsters were used. To prevent cross-contamination, donor and the sentinel cages were never opened at the same time, sentinel hamsters were not exposed to the same handling equipment as donors and after each sentinel the equipment was disinfected with either 70% ethanol or 5% Microchem.
Initially, transmission was studied assessing distance. Donor hamsters were infected intranasally as described above with 8 × 104 TCID50 SARS-CoV-2 (lineage A or B.1.1.7 variants). After 12 h, donor animals were placed into the donor cage and sentinels were placed into the sentinel cage (2:2). Airflow was generated between the cages from the donor to the sentinel cage at 30 changes h−1. Hamsters were cohoused at a 16.5, 106 or 200 cm distance. ALPH-dri bedding was used to avoid generation of dust particles. Oropharyngeal swabs were taken for donors at 1 d post-inoculation and for sentinels daily after exposure began. Swabs were collected in 1 ml DMEM with 200 U ml−1 penicillin and 200 µg ml−1 streptomycin. Exposure continued until respiratory shedding was confirmed in sentinels on three consecutive days. Then, donors were euthanized and sentinels were monitored until 14 d post-exposure for seroconversion.
Second, transmission was studied by assessing the duration of exposure. Donor hamsters were infected intranasally as described above with 8 × 104 TCID50 SARS-CoV-2. After 24 (1 d post-inoculation) or 72 h (3 d post-inoculation), donor animals were placed into the donor cage and sentinels were placed into the sentinel cage (2:2). Hamsters were cohoused at a 200 cm distance for 1 or 4 h at an airflow rate of 30 changes h−1. Oropharyngeal swabs were taken for donors on the day of exposure and for sentinels 3 d after exposure.
Variant competitiveness transmission chain
Four-to-six-week-old female and male Syrian hamsters were used. Donor hamsters (n = 8) were infected intranasally as described above with 1 × 102 TCID50 SARS-CoV-2 (as confirmed by back titration of the inoculum) at a 1:1 ratio of lineage A and B.1.1.7 mixture (54.7% B.1.1.7 as confirmed by full genome sequencing of the inoculum). After 12 h, donor animals were placed into the donor cage and sentinels (sentinels 1, n = 8) were placed into the sentinel cage (2:2) at a 16.5 cm distance at an airflow of 30 changes h−1. Hamsters were cohoused for 24 h. The following day, donor animals were rehoused into regular rodent caging and sentinels 1 were placed into the donor cage of new transmission set-ups. New sentinels (sentinels 2, n = 8) were placed into the sentinel cage (2:2) at a 16.5 cm distance at an airflow of 30 changes h−1. Hamsters were cohoused for 24 h. Then, sentinels 1 were rehoused into regular rodent caging and sentinels 2 were placed into the donor cage of new transmission set-ups. New sentinels (sentinels 3, n = 8) were placed into the sentinel cage (2:2) at a 16.5 cm distance at an airflow of 30 changes h−1. Hamsters were cohoused for 24 h. Then, both sentinels 2 and 3 were rehoused to regular rodent caging and monitored until 5 d post-exposure. Oropharyngeal swabs were taken for all animals at 2 d post-inoculation/post-exposure. All animals were euthanized at 5 d post-inoculation/post-exposure for collection of lung tissue.
Viral RNA detection
A total of 140 µl of swab sample was utilized for RNA extraction using the QIAamp Viral RNA Kit (QIAGEN) using the QIAcube HT automated system (QIAGEN) according to the manufacturer’s instructions with an elution volume of 150 µl. For tissues, RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions and eluted in 60 µl. Subgenomic viral RNA and genomic RNA was detected by RT–qPCR55. RNA was tested with TaqMan Fast Virus One-Step Master Mix using the QuantStudio 6 or 3 Flex Real-Time PCR System (Applied Biosystems). SARS-CoV-2 standards with known copy numbers were used to construct a standard curve and calculate copy numbers ml−1 or copy numbers g−1. Data were analysed using QuantStudio Software v.2.3.
Viral titration
Infectious virus in tissue and swab samples was determined as described previously56. Briefly, lung tissue samples were weighed then homogenized in 1 ml DMEM (2% FBS). VeroE6 cells were inoculated with tenfold serial dilutions of homogenate or swab media, incubated for 1 h at 37 °C and the first 2 dilutions washed twice with 2% DMEM. After 6 d, cells were scored for cytopathic effect. TCID50 ml−1 was calculated using the Spearman–Kärber method.
Serology
Serum samples were analysed as described previously57. Briefly, Nunc MaxiSorp plates were coated with 50 ng spike protein (generated in house) per well. Plates were incubated overnight at 4 °C. Plates were blocked with casein in PBS (Thermo Fisher Scientific) for 1 h at room temperature. Serum was diluted twofold in blocking buffer and samples (duplicate) were incubated for 1 h at room temperature. Secondary goat anti-hamster IgG Fc (horseradish peroxidase-conjugated, 1:2,500 dilution) spike-specific antibodies were used for detection and visualized with the KPL TMB 2-Component Microwell Peroxidase Substrate Kit (catalogue no. 5120-0047; SeraCare). The reaction was stopped with KPL stop solution (SeraCare) and plates were read at 450 nm. The threshold for positivity was calculated as the average plus 3× the s.d. of negative control hamster sera.
NGS of virus
For sequencing from swabs and lungs, total RNA was depleted of ribosomal RNA using the Ribo-Zero Gold rRNA Removal Kit (Illumina). Sequencing libraries were constructed using the KAPA RNA HyperPrep Kit according to the manufacturer’s protocol (Roche). To enrich for the SARS-CoV-2 sequence, libraries were hybridized to myBaits Expert Virus biotinylated oligonucleotide baits according to the manufacturer’s manual v.4.01 (Arbor Biosciences). Enriched libraries were sequenced on the Illumina MiSeq instrument as paired-end 2 × 150 bp reads. Raw FASTQ reads were trimmed of Illumina adaptor sequences using cutadapt v.1.1227 and then trimmed and filtered for quality using the FASTX-Toolkit v.0.0.14 (Hannon Lab, Cold Spring Harbor Laboratory). The remaining reads were mapped to the SARS-CoV-2 2019-nCoV/USA-WA1/2020 (MN985325.1) using Bowtie2 v.2.2.928 with the parameters --local--no-mixed -X 1500. PCR duplicates were removed using Picard MarkDuplicates v.2.26.9 (Broad Institute) and variants were called using GATK HaplotypeCaller v.4.1.2.029 with the parameter -ploidy 2. Variants were filtered for quality > 500 and depth > 20 using bcftools v.1.13-25-g41093eb+. We assessed the presence of N501Y, D614G and P681H and calculated the average to inform on the frequency of B.1.1.7 sequences in the sample.
Statistics and reproducibility
Power analysis was used to predetermine animal group size to allow statistical significance with 99% confidence intervals for assuming a fivefold difference in virus replication. Animals were randomly assigned to the experimental groups; investigators were not blinded to allocation during the experiments but were blinded during outcome assessment. Data distribution was assumed to be non-normal and non-parametric test were applied where appropriate. No data or animals were excluded from the analysis. Significance tests were performed as indicated where appropriate using Prism 8 (GraphPad Software). Statistical significance levels were determined as follows: NS, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data are available on request from the corresponding authors. All material requests should be sent to V.J.M., vincent.munster@nih.gov. Source data are provided with this paper.
References
Goldman, E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet Infect. Dis. 20, 892–893 (2020).
Pitol, A. K. & Julian, T. R. Community transmission of SARS-CoV-2 by fomites: risks and risk reduction strategies. Preprint at medRxiv https://doi.org/10.1101/2020.11.20.20220749 (2020).
Centers for Disease Control and Prevention. How COVID-19 Spreads (CDC, 2021); https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html
Boone, S. A. & Gerba, C. P. Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol. 73, 1687–1696 (2007).
Greenhalgh, T. et al. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 397, 1603–1605 (2021).
Coleman, K. K. et al. Viral load of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in respiratory aerosols emitted by patients with coronavirus disease 2019 (COVID-19) while breathing, talking, and singing. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciab691 (2021).
Hinds, W. C. in Aerosol Technology: Properties, Behavior, and Measurement of Airborne Particles 2nd edn, 53–70 (John & Wiley Sons, 1999).
Herfst, S. et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012).
Richard, M. et al. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 11, 766 (2020).
Yen, H.-L. et al. Hemagglutinin–neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc. Natl Acad. Sci. USA 108, 14264–14269 (2011).
Zhou, J. et al. Defining the sizes of airborne particles that mediate influenza transmission in ferrets. Proc. Natl Acad. Sci. USA 115, E2386–e2392 (2018).
Public Health England. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England. Technical Briefing 7 (Public Health England, 2021); https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/972247/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf
World Health Organization. Weekly Epidemiological Update-25 February 2021 (2021); https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update
Leung, N. H. L. et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 26, 676–680 (2020).
Chen, B. T., Cheng, Y. S. & Yeh, H. C. Performance of a TSI aerodynamic particle sizer. Aerosol Sci. Technol. 4, 89–97 (1985).
Sheely, M. L. Glycerol viscosity tables. Ind. Eng. Chem. 24, 1060–1064 (1932).
Laffeber, C., de Koning, K., Kanaar, R. & Lebbink, J. H. G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 433, 167058 (2021).
Port, J. R. et al. SARS-CoV-2 disease severity and transmission efficiency is increased for airborne but not fomite exposure in Syrian hamsters. Nat. Commun. 12, 4985 (2021).
Chan, J. F.-W. et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 71, 2428–2446 (2020).
Rosenke, K. et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 9, 2673–2684 (2020).
Suzuki, M. Environmental sampling for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during a coronavirus disease (COVID-19) outbreak aboard a commercial cruise ship. Preprint at medRxiv https://doi.org/10.1101/2020.05.02.20088567 (2020).
Lu, J. et al. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg. Infect. Dis. 26, 1628–1631 (2020).
Fennelly, K. P. Particle sizes of infectious aerosols: implications for infection control. Lancet Respir. Med. 8, 914–924 (2020).
Kutter, J. S. et al. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat. Commun. 12, 1653 (2021).
Katelaris, A. L. et al. Epidemiologic evidence for airborne transmission of SARS-CoV-2 during church singing, Australia, 2020. Emerg. Infect. Dis. 27, 1677–1680 (2021).
World Health Organization. Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions (WHO, 2020); https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions
Tellier, R., Li, Y., Cowling, B. J. & Tang, J. W. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect. Dis. 19, 101 (2019).
Sia, S. F. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020).
Bryche, B. et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav. Immun. 89, 579–586 (2020).
Richard, M. et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 11, 3496 (2020).
Milton, D. K. A Rosetta Stone for understanding infectious drops and aerosols. J. Pediatr. Infect. Dis. Soc. 9, 413–415 (2020).
Hou, Y. J. et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e14 (2020).
Gralton, J., Tovey, E., McLaws, M.-L. & Rawlinson, W. D. The role of particle size in aerosolised pathogen transmission: a review. J. Infect. 62, 1–13 (2011).
Yinda, C. K. et al. Prior aerosol infection with lineage A SARS-CoV-2 variant protects hamsters from disease, but not reinfection with B.1.351 SARS-CoV-2 variant. Emerg. Microbes Infect. 10, 1284–1292 (2021).
Volz, E. et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021).
Lindstrøm, J. C. et al. Increased transmissibility of the B.1.1.7 SARS-CoV-2 variant: evidence from contact tracing data in Oslo, January to February 2021. Preprint at medRxiv https://doi.org/10.1101/2021.03.29.21254122 (2021).
Adenaiye, O. O. et al. Infectious SARS-CoV-2 in exhaled aerosols and efficacy of masks during early mild infection. Clin. Infect. Dis., https://doi.org/10.1093/cid/ciab797 (2021).
Mohandas, S. et al. Comparison of SARS-CoV-2 variants of concern 202012/01 (U.K. variant) and D614G variant transmission by different routes in Syrian hamsters. Vector Borne Zoonotic Dis. 21, 638–641 (2021).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020).
Zhou, B. et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592, 122–127 (2021).
Ulrich, L. et al. Enhanced fitness of SARS-CoV-2 variant of concern B.1.1.7, but not B.1.351, in animal models. Preprint at bioRxiv https://doi.org/10.1101/2021.06.28.450190 (2021).
Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020).
Lauring, A. S. & Hodcroft, E. B. Genetic variants of SARS-CoV-2—What do they mean? JAMA 325, 529–531 (2021).
Koster, F. et al. Exhaled aerosol transmission of pandemic and seasonal H1N1 influenza viruses in the ferret. PLoS ONE 7, e33118 (2012).
Morawska, L. et al. How can airborne transmission of COVID-19 indoors be minimised? Environ. Int. 142, 105832 (2020).
Nardell, E. A. & Nathavitharana, R. R. Airborne spread of SARS-CoV-2 and a potential role for air disinfection. JAMA 324, 141–142 (2020).
Chu, D. K. et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet 395, 1973–1987 (2020).
Fukushi, S. et al. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 86, 2269–2274 (2005).
Kawase, M., Shirato, K., Matsuyama, S. & Taguchi, F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 83, 712–721 (2009).
Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020).
Takada, A. et al. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl Acad. Sci. USA 94, 14764–14769 (1997).
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020).
van Doremalen, N. et al. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antivir. Res. 143, 30–37 (2017).
Yinda, C. K. et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 17, e1009195 (2021).
Acknowledgements
We thank E. de Wit, B. Williamson, N. Thornburg, S. Tong, S. Rashid, R. Mukul, K. Stemple, C. Martens, K. Barbian, S. Ricklefs, S. Anzick, R. Perry, T. Jones, R. Stehlik, S. Cooley and S. Sarchette and the animal caretakers for their assistance during the study. The following reagent was obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): SARS-Related Coronavirus 2, Isolate hCoV-19/England/204820464/2020, NR282 54000, contributed by B. Hallis. This work was supported by the Intramural Research Program of NIAID, NIH (no. 1ZIAAI001179-01). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.R.P., C.K.Y. and V.J.M. designed the study. J.R.P., C.K.Y., R.J.F., V.A.A., M.G.H., J.E.S. and N.V.D. performed the experiments. J.R.P., C.K.Y. and C.S. analysed the results. J.R.P., C.K.Y. and V.J.M. wrote the manuscript. All coauthors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1–6.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Supplementary Fig. 2
Statistical source data.
Source Data Supplementary Fig. 3
Statistical source data.
Rights and permissions
About this article
Cite this article
Port, J.R., Yinda, C.K., Avanzato, V.A. et al. Increased small particle aerosol transmission of B.1.1.7 compared with SARS-CoV-2 lineage A in vivo. Nat Microbiol 7, 213–223 (2022). https://doi.org/10.1038/s41564-021-01047-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-01047-y
This article is cited by
-
Augmentation of Omicron BA.1 pathogenicity in hamsters using intratracheal inoculation
npj Viruses (2024)
-
Airborne transmission efficiency of SARS-CoV-2 in Syrian hamsters is not influenced by environmental conditions
npj Viruses (2024)
-
The evolution of SARS-CoV-2
Nature Reviews Microbiology (2023)
-
Infection- or AZD1222 vaccine-mediated immunity reduces SARS-CoV-2 transmission but increases Omicron competitiveness in hamsters
Nature Communications (2023)
-
A frontal air intake may improve the natural ventilation in urban buses
Scientific Reports (2022)