Abstract
Synthesis of septal peptidoglycan (sPG) is crucial for bacterial cell division. FtsW, an indispensable component of the cell division machinery in all walled bacterial species, was recently identified in vitro as a peptidoglycan glycosyltransferase (PGTase). Despite its importance, the septal PGTase activity of FtsW has not been demonstrated in vivo. How its activity is spatiotemporally regulated in vivo has also remained elusive. Here, we confirmed FtsW as an essential septum-specific PGTase in vivo using an N-acetylmuramic acid analogue incorporation assay. Next, using single-molecule tracking coupled with genetic manipulations, we identified two populations of processively moving FtsW molecules: a fast-moving population correlated with the treadmilling dynamics of the essential cytoskeletal FtsZ protein and a slow-moving population dependent on active sPG synthesis. We further identified that FtsN, a potential sPG synthesis activator, plays an important role in promoting the slow-moving population. Our results suggest a two-track model, in which inactive sPG synthases follow the ‘Z-track’ to be distributed along the septum and FtsN promotes their release from the Z-track to become active in sPG synthesis on the slow ‘sPG-track’. This model provides a mechanistic framework for the spatiotemporal coordination of sPG synthesis in bacterial cell division.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data, plasmids and E. coli strains that support the findings of this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
The in-house developed image analysis scripts are available in the GitHub repository: https://github.com/XiaoLabJHU/SMT_Unwrapping.
References
Du, S. & Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 (2017).
McQuillen, R. & Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-Ring. Annu. Rev. Biophys. 49, 309–341 (2020).
Yang, X. et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017).
Bisson-Filho, A. W. et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017).
Taguchi, A. et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019).
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016).
Butler, E. K., Davis, R. M., Bari, V., Nicholson, P. A. & Ruiz, N. Structure–function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J. Bacteriol. 195, 4639–4649 (2013).
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Sjodt, M. et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 556, 118–121 (2018).
Karlin, A. & Akabas, M. H. Substituted-cysteine accessibility method. Methods Enzymol. 293, 123–145 (1998).
Liang, H. et al. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat. Commun. 8, 15015 (2017).
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. 40, 2004–2021 (2001).
Sham, L.-T. et al. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 (2014).
Egan, A. J. F. & Vollmer, W. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277, 8–28 (2012).
Leclercq, S. et al. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 7, 43306 (2017).
Fraipont, C. et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 (2010).
Ovchinnikov, S. et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife 4, e09248 (2015).
Karimova, G., Dautin, N. & Ladant, D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187, 2233–2243 (2005).
McCausland, J. W., Yang, X., Lyu, Z. & Söderström, B. W. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Preprint at bioRxiv https://doi.org/10.1101/857813 (2019).
Perez, A. J. et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA 116, 3211–3220 (2019).
Söderström, B., Chan, H., Shilling, P. J., Skoglund, U. & Daley, D. O. Spatial separation of FtsZ and FtsN during cell division. Mol. Microbiol. 107, 387–401 (2017).
Li, Z., Trimble, M. J., Brun, Y. V. & Jensen, G. J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708 (2007).
Squyres, G. R., Holmes, M. J., Barger, S. R. & Pennycook, B. R. Dynamics of bacterial cell division: Z ring condensation is essential for cytokinesis. Preprint at bioRxiv https://doi.org/10.1101/2020.06.30.180737 (2020).
Coltharp, C., Buss, J., Plumer, T. M. & Xiao, J. Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl Acad. Sci. USA 113, E1044–E1053 (2016).
Park, K. T., Du, S. & Lutkenhaus, J. Essential role for FtsL in activation of septal peptidoglycan synthesis. mBio 11, e0301220 (2020).
Kocaoglu, O. & Carlson, E. E. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob. Agents Chemother. 59, 2785–2790 (2015).
Özbaykal, G., Wollrab, E., Simon, F. & Vigouroux, A. The transpeptidase PBP2 governs initial localization and activity of the major cell-wall synthesis machinery in E. coli. eLife 9, e50629 (2020).
Lee, T. K. et al. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proc. Natl Acad. Sci. USA 111, 4554–4559 (2014).
Rojas, E., Theriot, J. A. & Huang, K. C. Response of Escherichia coli growth rate to osmotic shock. Proc. Natl Acad. Sci. USA 111, 7807–7812 (2014).
Silver, L. L. Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7, a025262 (2017).
Cho, H., Uehara, T. & Bernhardt, T. G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014).
Liu, B., Persons, L., Lee, L. & de Boer, P. A. J. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945–970 (2015).
Tsang, M.-J. & Bernhardt, T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015).
Tarry, M. et al. The Escherichia coli cell division protein and model Tat substrate SufI (FtsP) localizes to the septal ring and has a multicopper oxidase-like structure. J. Mol. Biol. 386, 504–519 (2008).
Vollmer, W. & Seligman, S. J. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18, 59–66 (2010).
Burman, L. G. & Park, J. T. Molecular model for elongation of the murein sacculus of Escherichia coli. Proc. Natl Acad. Sci. USA 81, 1844–1848 (1984).
Neidhardt, F. C., Bloch, P. L. & Smith, D. F. Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974).
Buss, J. et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128 (2015).
Glascock, C. B. & Weickert, M. J. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene 223, 221–231 (1998).
Moore, D. A., Whatley, Z. N., Joshi, C. P., Osawa, M. & Erickson, H. P. Probing for binding regions of the FtsZ protein surface through site-directed insertions: discovery of fully functional FtsZ-fluorescent proteins. J. Bacteriol. 199, 18 (2016).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016).
Sambrook, J. & Russell, D. W. The Condensed Protocols from ‘Molecular Cloning: A Laboratory Manual’ (Cold Spring Harbor Laboratory Press, 2006).
Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P. & Kushner, S. R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171, 4617–4622 (1989).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (2008).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Sbalzarini, I. F. & Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151, 182–195 (2005).
Lyu, Z., Coltharp, C., Yang, X. & Xiao, J. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional superresolution imaging. Biopolymers 105, 725–734 (2016).
Buss, J. et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120 (2013).
Kusumi, A., Sako, Y. & Yamamoto, M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Methods Enzymol. 65, 2021–2040 (1993).
Acknowledgements
We thank other laboratory members in the Xiao and de Boer laboratories for helpful discussions and technical assistance. We also thank G. Hauk for sharing plasmids and the CRISPR-Cas9/λ-red recombineering cloning method, D. S. Weiss for strain EC1908, plasmid pDSW406, anti-FtsN serum and helpful suggestions on FtsW immunoblotting, T. Bernhardt for strain HC532 and plasmid pHC808, C. Hale for plasmid pCH650, E. Goley for help on cell growth measurement and R. Tsien for the TagRFP-T construct. This work was supported by NIH U01CA221230 (to C.L.G.), Pew Biomedical Scholar (Pew Foundation to C.L.G.), NIH T32GM133395A (to K.E.D.), NIH GM57059 (to P.d.B.), NIH R01GM086447 and R35GM136436 (to J.X.), GM125656 (subcontract to J.X.), NSF EAGER award MCB-1019000 (to J.X.) and a Hamilton Smith Innovative Research Award (to J.X.).
Author information
Authors and Affiliations
Contributions
X.Y., P.d.B. and J.X. conceptualized the experiments. X.Y. performed single-molecule tracking experiments and analysed the data. R.M., H.L., K.E.D. and C.C.S. optimized and performed cell wall labelling experiments. X.Y. and R.M. analysed the cell wall labelling data. P.P.-M. isolated the FtsW and FtsI SF mutants and performed genetic and phenotypic experiments. X.Y. and A.D.L.C. screened FtsW single cysteine mutations. J.W.M. and A.D.L.C. performed phenotypic experiments of other ftsW mutation strains. X.Y., P.d.B. and J.X. wrote the original draft. X.Y., R.M., Z.L., J.W.M., H.L., K.E.D., P.d.B. and J.X. reviewed and edited the manuscript. X.Y., C.L.G, P.d.B. and J.X. supervised the study. Funding was acquired by C.L.G, P.d.B. and J.X.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single Cys residue substitutions in FtsW alter its function and cell morphology.
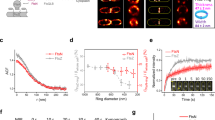
a, Homology structure of FtsW based on the structure of RodA (PDB: 6BAR)9 using Phyre 2.045. Mutated residues are labelled and colour-coded according to their sensitivity to MSTES. Green: MTSES does not affect cell growth or morphology (S131C, L276C, and L307C). Orange: MTSES significantly slows down cell division and leads to elongated cells (S136C, L198C, L288C, and A294C). Red: MTSES blocks cell division completely and leads to long, chaining cells (A301C, I302C, I303C, and L367C). Magenta: Cysteine mutation causes other cell division defects such as abnormal septa and cell poles even in the absence of MTSES (L268C). b, Time-lapse images of JXY559 (ftswI302C) cells in M9 medium without (top panel) and with (bottom panel) MTSES. MTSES was added to the cell in the bottom panel 30 min before the first image (Supplementary Videos 1 and 2). Arrowhead marks a cell constriction that initiated early on but failed to complete. Scale bars: 2 µm. c, Integrated intensity of septal NAM labelling of different strains in the absence or presence of MTSES. Error bars represent S.E.M of three independent experiments with the same imaging condition.
Extended Data Fig. 2 Representative septal FtsW-RFP trajectories are segmented and fit with line to extract directional-moving speeds.
Raw trajectories of single FtsW-RFP molecules along the septal circumference are demonstrated as orange lines. Gary, light-yellow, and light-blue shades indicate segments of stationary, low-moving, and fast-moving states respectively. Corresponding speeds, obtained by linear fitting, are shown as solid line and labelled in each sub-figure. Supplementary videos 6 to 9 correspond to trajectories a to d.
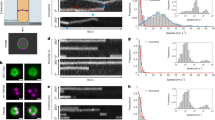
Extended Data Fig. 3 FtsW-RFP molecules in stationary state display confined diffusion.
a, Example of an FtsW-RFP molecule (arrowhead) remaining within a small region of the septum. As indicated by the kymograph, this molecule showed little observable directional movement for at least 80 seconds. b, Unwrapped and decomposed 1d-trajectory of the FtsW-RFP molecule along the long (grey) and short (orange) axis. c, 1d-MSD curves (short axis) of 204 ‘stationary’ molecules in live BW25113 WT cells growing in M9-glucose medium without (blue) or with 1 µg/mL Aztreonam (dark green), and in cells grown without Aztreonam upon paraformaldehyde fixation (grey). The BW25113 curve (blue) is fit by Kusumi equation52 with D = 0.0007 ± 0.0002 µm2/s, L = 95 ± 6 nm. Note that Aztreonam treatment causes confinement of ‘stationary’ FtsW-RFP molecules to smaller areas, similar to what is observed in fixed cells. d, Percentage of FtsW-RFP molecules in stationary state in the presence (blue bars) and absence (orange bars) of drug to inhibit sPG synthesis (MTSES for FtsWI302C and PBP1BS247C, Aztreonam for FtsI, and Fosfomycin for precursor synthesis). e, Mean dwell time of FtsW-RFP molecules staying in stationary state in the presence (blue dots) and absence (orange dots) of drug to inhibit sPG synthesis. c and d. Data are presented as mean ± s.e.m. where the s.e.m. is estimated by bootstrapping (For details regarding statistics and box plot definitions see the ‘Statistics’ subheading in ‘Methods’). e, Data are presented as mean ± s.e.m.. For the sample size of each point see Supplementary Table 6.
Extended Data Fig. 4 Speed distributions of processive moving RFP-FtsI molecules and FtsZ treadmilling under different conditions.
Speed distribution (bars) of all RFP-FtsI molecules in ftsZ wt (a). GTPase mutant (ftsZE250A) (b), and ftsB SF mutant (ftsBE56A) (c) strains. The CDF fit curves of fast- (blue solid) and slow- (red solid) moving population are overlaid with the bar graph. The black dash curves indicate the overall fitting. FtsZ treadmilling speed distribution in different FtsZ GTPase mutant strains (d) (Data from3), in superfission mutant strains and different growth media (e), and under different drug treatment conditions (f). The average speeds under all conditions are given in Supplementary Tables 4 and 6.
Extended Data Fig. 5 Superfission (SF) mutations in ftsW or ftsI allow for growth and division in cells with diminished or no FtsN function.
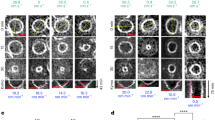
a, Differential interference contrast (DIC) cell images of strains TB28 (wt), CH34/pBL145 (∆ftsN / cIts PλR::ftsN1–90), PM17 (ftsWE289G), PM18 (ftsWE289G ∆ftsN), and PM6 (ftsIR167S). Cells were grown overnight in LB medium at 37 °C (CH34/pBL145) or 30 °C (all other sttrains), diluted to OD600 = 0.02 in LB, and grown to OD600 = 0.6–0.7 at 30 oC (causing depletion of FtsN in the CH34/pBL145 cells shown). Scale bars: 4 µm. b, DIC cell images of strains TB77 (ftsNslm117), and PM4 (ftsNslm117 ftsIR167S). Overnight cultures were diluted to OD600 = 0.05 in M9-glucose medium and grown to OD600 = 0.5–0.6 at 30 oC. Note that the ftsNslm117 allele corresponds to an EZTnKan-2 transposon insertion in codon 119 of ftsN, leading to a pronounced, but non-lethal, cell constriction defect. Also note that the ftsIR167S allele in strain PM4 largely overcomes this defect. Scale bars: 8 µm. Representative micrographs were from three independent experiments. For the number of cells and characterization of cell morphology, see Supplementary Table 7.
Extended Data Fig. 6 The constriction rate of FtsI SF strain (TB28, ftsIR167S) is correlated with the cell growth rate.
a, Growth curves of PM6 in M9-acetate (magenta), M9-glucose (orange), and EZRDM (olive). Data are presented as mean ± s.d. from three independent experiments. b, Representative kymographs of the septum closure progress in PM6 cells growing in the indicated media, as probed by mNeonGreen-ZapA fluorescence. Representative micrographs were from two independent experiments. Scale bars: 200 nm. c, Relationship between the cell growth rate (Vg, estimated from a) and the constriction rate (Vc, estimated from kymographs as in b). The boxes indicate the ± SEM, whiskers the 10–90 percentiles, the midline indicates median, the square indicates mean. b,c, For the sample size of condition see Supplementary Table 8.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Table 1–9, Discussion, captions for Videos 1–9 and references.
Supplementary Video 1
Phase contrast time-lapse video of JXY559 (BW25113 FtsWI302C) cells growing on 3% agarose gel pad with M9-glucose minimum medium in the presence of MTSES at room temperature. The video was recorded every ~3.5 min. Scale bar, 5 μm.
Supplementary Video 2
Phase contrast time-lapse video of JXY559 (BW25113 FtsWI302C) cells growing on 3% agarose gel pad with M9-glucose minimum medium at room temperature. The gel pad was supplemented with 0.1 mM MTSES. The video was recorded every ~3.5 min. Scale bar, 5 μm.
Supplementary Video 3
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Extended Data Fig. 3a. Scale bar, 0.5 μm.
Supplementary Video 4
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Fig. 2a. Scale bar, 0.5 μm.
Supplementary Video 5
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Fig. 2b. Scale bar, 0.5 μm.
Supplementary Video 6
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Extended Data Fig. 2a. Scale bar, 0.5 μm.
Supplementary Video 7
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Extended Data Fig. 2b. Scale bar, 0.5 μm.
Supplementary Video 8
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Extended Data Fig. 2c. Scale bar, 0.5 μm.
Supplementary Video 9
Epifluorescence time-lapse imaging of single FtsW-RFP molecules in M9-glucose, corresponding to Extended Data Fig. 2d. Scale bar, 0.5 μm.
Supplementary Data 1
Source data for Supplementary Figs. 1–6.
Supplementary Data 2
Unmodified immunoblot image for Fig. 1.
Supplementary Data 3
Unmodified immunoblot image for Fig. 6.
Source data
Source Data Fig. 1
Source data.
Source Data Fig. 2
Source data.
Source Data Fig. 3
Source data.
Source Data Fig. 4
Source data.
Source Data Fig. 5
Source data.
Source Data Extended Data Fig. 1
Source data.
Source Data Extended Data Fig. 2
Source data.
Source Data Extended Data Fig. 3
Source data.
Source Data Extended Data Fig. 4
Source data.
Source Data Extended Data Fig. 6
Source data.
Rights and permissions
About this article
Cite this article
Yang, X., McQuillen, R., Lyu, Z. et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat Microbiol 6, 584–593 (2021). https://doi.org/10.1038/s41564-020-00853-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-00853-0
This article is cited by
-
Insights into the assembly and regulation of the bacterial divisome
Nature Reviews Microbiology (2024)
-
Cell constriction requires processive septal peptidoglycan synthase movement independent of FtsZ treadmilling in Staphylococcus aureus
Nature Microbiology (2024)
-
Peptidoglycan synthesis drives a single population of septal cell wall synthases during division in Bacillus subtilis
Nature Microbiology (2024)
-
Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism
Nature Communications (2023)
-
Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli
Nature Microbiology (2022)