Abstract
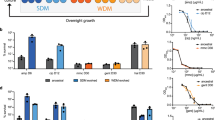
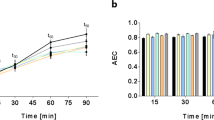
Growth rate and metabolic state of bacteria have been separately shown to affect antibiotic efficacy1,2,3. However, the two are interrelated as bacterial growth inherently imposes a metabolic burden4; thus, determining individual contributions from each is challenging5,6. Indeed, faster growth is often correlated with increased antibiotic efficacy7,8; however, the concurrent role of metabolism in that relationship has not been well characterized. As a result, a clear understanding of the interdependence between growth and metabolism, and their implications for antibiotic efficacy, are lacking9. Here, we measured growth and metabolism in parallel across a broad range of coupled and uncoupled conditions to determine their relative contribution to antibiotic lethality. We show that when growth and metabolism are uncoupled, antibiotic lethality uniformly depends on the bacterial metabolic state at the time of treatment, rather than growth rate. We further reveal a critical metabolic threshold below which antibiotic lethality is negligible. These findings were general for a wide range of conditions, including nine representative bactericidal drugs and a diverse range of Gram-positive and Gram-negative species (Escherichia coli, Acinetobacter baumannii and Staphylococcus aureus). This study provides a cohesive metabolic-dependent basis for antibiotic-mediated cell death, with implications for current treatment strategies and future drug development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
Zampieri, M., Zimmermann, M., Claassen, M. & Sauer, U. Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep. 19, 1214–1228 (2017).
Yang, J. H., Bening, S. C. & Collins, J. J. Antibiotic efficacy—context matters. Curr. Opin. Microbiol. 39, 73–80 (2017).
Lee, A. J. et al. Robust, linear correlations between growth rates and β-lactam–mediated lysis rates. Proc. Natl Acad. Sci. USA 115, 4069–4074 (2018).
Lipson, D. A. The complex relationship between microbial growth rate and yield and its implications for ecosystem processes. Front. Microbiol. 6, 615 (2015).
Brown, M. R., Collier, P. J. & Gilbert, P. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 34, 1623–1628 (1990).
Russell, J. B. The energy spilling reactions of bacteria and other organisms. J. Mol. Microbiol. Biotechnol. 13, 1–11 (2007).
Greulich, P., Scott, M., Evans, M. R. & Allen, R. J. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 11, 796 (2015).
Haugan, M. S., Løbner-Olesen, A. & Frimodt-Møller, N. Comparative activity of ceftriaxone, ciprofloxacin, and gentamicin as a function of bacterial growth rate probed by Escherichia coli chromosome replication in the mouse peritonitis model. Antimicrob. Agents Chemother. 63, e02133-18 (2019).
Fisher, J. F. & Mobashery, S. Endless resistance. Endless antibiotics? MedChemComm 7, 37–49 (2016).
Low, E. W. & Chase, H. A. Reducing production of excess biomass during wastewater treatment. Water Res. 33, 1119–1132 (1999).
Russell, J. B. & Cook, G. M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59, 48–62 (1995).
Molenaar, D., van Berlo, R., de Ridder, D. & Teusink, B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5, 323 (2009).
Erickson, D. W. et al. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 551, 119–123 (2017).
Manzoni, S., Taylor, P., Richter, A., Porporato, A. & Ågren, G. I. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 196, 79–91 (2012).
Lahtvee, P.-J. et al. Multi-omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microb. Cell Fact. 10, 12 (2011).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Waschina, S., Souza, G. D., Kost, C. & Kaleta, C. Metabolic network architecture and carbon source determine metabolite production costs. FEBS J. 283, 2149–2163 (2016).
Akashi, H. & Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA 99, 3695–3700 (2002).
Gschaedler, A. & Boudrant, J. Amino acid utilization during batch and continuous cultures of Escherichia coli on a semi-synthetic medium. J. Biotechnol. 37, 235–251 (1994).
Orth, J. D. et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol. Syst. Biol. 7, 535 (2011).
Holms, H. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19, 85–116 (1996).
Kempes, C. P. et al. Drivers of bacterial maintenance and minimal energy requirements. Front. Microbiol. 8, 31 (2017).
Shan, Y. et al. ATP-dependent persister formation in Escherichia coli. mBio 8, 02267-16 (2017).
Chang, D.-E., Smalley, D. J. & Conway, T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45, 289–306 (2002).
Rampersad, S. N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 12, 12347–12360 (2012).
Yang, J. H. et al. A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell 177, 1649–1661 (2019).
Lobritz, M. A. et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl Acad. Sci. USA 112, 8173–8180 (2015).
Dwyer, D. J. et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl Acad. Sci. USA 111, E2100–E2109 (2014).
Wright, N. E. & Gilbert, P. Influence of specific growth rate and nutrient limitation upon the sensitivity of Escherichia coli towards chlorhexidine diacetate. J. Appl. Bacteriol. 62, 309–314 (1987).
Lopatkin, A. J. et al. Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1, 16044 (2016).
Lopatkin, A. J. et al. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 8, 1689 (2017).
Schellenberger, J. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307 (2011).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Theodosiou, E., Frick, O., Bühler, B. & Schmid, A. Metabolic network capacity of Escherichia coli for Krebs cycle-dependent proline hydroxylation. Microb. Cell Fact. 14, 108 (2015).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Gottschalk, G. Bacterial Metabolism (Springer, 1979).
Acknowledgements
We thank D. Hung from the Broad Institute for providing the bacterial pathogens used in this study; J. K. Srimani, R. P. Smith and S. Bening for input on data interpretation and manuscript editing. This work was supported by the Defence Threat Reduction Agency (grant number HDTRA1-15-1-0051), the National Institutes of Health (grant number K99GM118907), the Banting Postdoctoral Fellowship Program (to J.M.S.; grant number 393360), the Broad Institute of MIT and Harvard, and a generous gift from A. Bekenstein and J. Bekenstein.
Author information
Authors and Affiliations
Contributions
A.J.L. conceived the research, designed and carried out experiments and data analysis, developed the simplified model, interpreted data and wrote the manuscript. J.M.S. assisted with data acquisition and interpretation and manuscript editing. E.J.Z. assisted with data acquisition and manuscript editing. J.H.Y. assisted with metabolic modelling, data interpretation and manuscript editing. M.K.T. assisted with data acquisition and interpretation and manuscript editing. L.Y. assisted with data interpretation, model development and manuscript editing. J.J.C. conceived the research and assisted with data interpretation and manuscript editing.
Corresponding author
Ethics declarations
Competing interests
J.J.C. is scientific co-founder and Scientific Advisory Board chair of EnBiotix, an antibiotic drug discovery company.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary Figs. 1–11, Supplementary Tables 1–8 and Supplementary References.
Rights and permissions
About this article
Cite this article
Lopatkin, A.J., Stokes, J.M., Zheng, E.J. et al. Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat Microbiol 4, 2109–2117 (2019). https://doi.org/10.1038/s41564-019-0536-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0536-0
This article is cited by
-
Rapid Mycobacterium abscessus antimicrobial susceptibility testing based on antibiotic treatment response mapping via Raman Microspectroscopy
Annals of Clinical Microbiology and Antimicrobials (2023)
-
Metabolic genes on conjugative plasmids are highly prevalent in Escherichia coli and can protect against antibiotic treatment
The ISME Journal (2023)
-
The non-attached biofilm aggregate
Communications Biology (2023)
-
Profiling cell envelope-antibiotic interactions reveals vulnerabilities to β-lactams in a multidrug-resistant bacterium
Nature Communications (2023)
-
Wet-dry cycles protect surface-colonizing bacteria from major antibiotic classes
The ISME Journal (2022)