Abstract
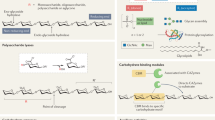
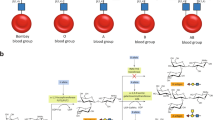
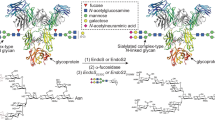
Access to efficient enzymes that can convert A and B type red blood cells to ‘universal’ donor O would greatly increase the supply of blood for transfusions. Here we report the functional metagenomic screening of the human gut microbiome for enzymes that can remove the cognate A and B type sugar antigens. Among the genes encoded in our library of 19,500 expressed fosmids bearing gut bacterial DNA, we identify an enzyme pair from the obligate anaerobe Flavonifractor plautii that work in concert to efficiently convert the A antigen to the H antigen of O type blood, via a galactosamine intermediate. The X-ray structure of the N-acetylgalactosamine deacetylase reveals the active site and mechanism of the founding member of an esterase family. The galactosaminidase expands activities within the CAZy family GH36. Their ability to completely convert A to O of the same rhesus type at very low enzyme concentrations in whole blood will simplify their incorporation into blood transfusion practice, broadening blood supply.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets generated and/or analysed during this study are available from the corresponding author on reasonable request. Structure datasets generated during the current study are available in the PDB repository under accession numbers 6N1A and 6N1B. The protein CspGH36 identified during the current study is deposited in GenBank under the accession code MK500922.
References
Daniels, G. The molecular definition of red cell antigens. ISBT Sci. Ser. 5, 300–302 (2010).
Garratty, G. Modulating the red cell membrane to produce universal/stealth donor red cells suitable for transfusion. Vox Sang. 94, 87–95 (2008).
Goldstein, J., Siviglia, G., Hurst, R., Lenny, L. & Reich, L. Group-B erythrocytes enzymatically converted to Group-O survive normally in A, B, and O individuals. Science 215, 168–170 (1982).
Kruskall, M. S. et al. Transfusion to blood group A and O patients of group B RBCs that have been enzymatically converted to group O. Transfusion 40, 1290–1298 (2000).
Clausen, H. & Hakomori, S. Abh and related histo-blood group antigens - immunochemical differences in carrier isotypes and their distribution. Vox Sang. 56, 1–20 (1989).
Liu, Q. P. et al. Bacterial glycosidases for the production of universal red blood cells. Nat. Biotechnol. 25, 454–464 (2007).
Anderson, K. M. et al. A clostridial endo-beta-galactosidase that cleaves both blood group A and B glycotopes. J. Biol. Chem. 280, 7720–7728 (2005).
Kwan, D. H. et al. Toward efficient enzymes for the generation of universal blood through structure-guided directed evolution. J. Am. Chem. Soc. 137, 5695–5705 (2015).
Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. R. 68, 669–685 (2004).
Amann, R. I. et al. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl. Environ. Microbiol. 56, 1919–1925 (1990).
Tailford, L. E., Crost, E. H., Kavanaugh, D. & Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81 (2015).
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014).
Konwar, K. M. et al. MetaPathwaysv2.5: quantitative functional, taxonomic and usability improvements. Bioinformatics 31, 3345–3347 (2015).
Li, H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015).
Yip, V. L. Y. et al. An unusual mechanism of glycoside hydrolysis involving redox and elimination steps by a family 4 beta-glycosidase from Thermotoga maritima. J. Am. Chem. Soc. 126, 8354–8355 (2004).
Chapanian, R. et al. Enhancement of biological reactions on cell surfaces via macromolecular crowding. Nat. Commun. 5, 4683 (2014).
Comfort, D. A. et al. Biochemical analysis of Thermotoga maritima GH36 alpha-galactosidase (TmGalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry 46, 3319–3330 (2007).
Fredslund, F. et al. Crystal structure of alpha-galactosidase from Lactobacillus acidophilus NCFM: insight into tetramer formation and substrate binding. J. Mol. Biol. 412, 466–480 (2011).
Calcutt, M. J., Hsieh, H. Y., Chapman, L. F. & Smith, D. S. Identification, molecular cloning and expression of an alpha-N-acetylgalactosaminidase gene from Clostridium perfringens. FEMS Microbiol. Lett. 214, 77–80 (2002).
Gerbal, A., Maslet, C. & Salmon, C. Immunological aspects of the acquired B antigen. Vox Sang. 28, 398–403 (1975).
Judd, W. J. & Annesley, T. M. The acquired-B phenomenon. Transfus. Med. Rev. 10, 111–117 (1996).
Marcus, D. M., Kabat, E. A. & Schiffman, G. Immunochemical studies on blood groups. XXXI. Destruction of blood group a activity by an enzyme from Clostridium tertium which deacetylates N-acetylgalactosamine in intact blood group substances. Biochemistry 3, 437–443 (1964).
Yamamoto, H. & Iseki, S. Development of H-specificity in A substance by A-decomposing enzyme from Clostridium tertium A. P. Jpn Acad. 44, 263 (1968).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Blair, D. E., Schuttelkopf, A. W., MacRae, J. I. & van Aalten, D. M. F. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl Acad. Sci. USA 102, 15429–15434 (2005).
Ficko-Blean, E. & Boraston, A. B. The interaction of a carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor. J. Biol. Chem. 281, 37748–37757 (2006).
Cohen, M., Hurtado-Ziola, N. & Varki, A. ABO blood group glycans modulate sialic acid recognition on erythrocytes. Glycobiology 19, 1349–1349 (2009).
Hyono, A. et al. Impacts of papain and neuraminidase enzyme treatment on electrohydrodynamics and IgG-mediated agglutination of type A red blood cells. Langmuir 25, 10873–10885 (2009).
Varki, A. et al. Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 (2015).
Kwan, D. H., Ernst, S., Kötzler, M. P. & Withers, S. G. Chemoenzymatic synthesis of a type 2 blood group a tetrasaccharide and development of high-throughput assays enables a platform for screening blood group antigen-cleaving enzymes. Glycobiology 25, 806–811 (2015).
Armstrong, Z., Rahfeld, P. & Withers, S. G. Discovery of new glycosidases from metagenomic libraries. Methods Enzymol. Chem. Glycobiol. A 597, 3–23 (2017).
Lee, S. & Hallam, S. J. Extraction of high molecular weight genomic DNA from soils and sediments. J. Vis. Exp. 33, e1569 (2009).
Lee, E., Chuang, H. Y., Kim, J. W., Ideker, T. & Lee, D. Inferring pathway activity toward precise disease classification. PloS Comput. Biol. 4, e1000217 (2008).
Jeong, J. K. et al. Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface beta-galactosidase with specific hydrolysis activity for the gal beta 1-3GlcNAc moiety of oligosaccharides. J. Bacteriol. 191, 3011–3023 (2009).
Singh, A. K. et al. Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. PLoS Pathog. 10, e1004364 (2014).
Katayama, T. et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186, 4885–4893 (2004).
Williams, S. J. & Withers, S. G. Glycosynthases: mutant glycosidases for glycoside synthesis. Aust. J. Chem. 55, 3–12 (2002).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv https://arxiv.org/abs/1303.3997 (2013).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Treangen, T. J., Sommer, D. D., Angly, F. E., Koren, S. & Pop, M. Next generation sequence assembly with AMOS. Curr. Protoc. Bioinformatics 33, 11.8.1–11.8.18 (2011).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Studier, F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Klock, H. E., Koesema, E. J., Knuth, M. W. & Lesley, S. A. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994 (2008).
Palmier, M. O. & Van Doren, S. R. Rapid determination of enzyme kinetics from fluorescence: overcoming the inner filter effect. Anal. Biochem. 371, 43–51 (2007).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Skubak, P. & Pannu, N. S. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013).
Potterton, L. et al. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D 74, 68–84 (2018).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Zheng, L., Baumann, U. & Reymond, J. L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115 (2004).
Vocadlo, D. J., Wicki, J., Rupitz, K. & Withers, S. G. Mechanism of Thermoanaerobacterium saccharolyticum ss-xylosidase: kinetic studies. Biochemistry 41, 9727–9735 (2002).
Jones, D. R. et al. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol. Biofuels 11, 27 (2018).
Yin, Y. B. et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451 (2012).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 (2008).
Eddy, S. R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Matsen, F. A., Hoffman, N. G., Gallagher, A. & Stamatakis, A. A format for phylogenetic placements. PLoS ONE 7, e31009 (2012).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Acknowledgements
We thank H. Brumer for the use of his Dionex high-performance liquid chromatography system, H.-M. Chen for the synthesis of Gal-α-MU and GalNAc-α-MU and T. Lowary for a set of α-GalNAc-containing oligosaccharide substrates. We acknowledge the participation of the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grant P41 GM103694). We thank L. Worrall and N. Cavaney for collecting the X-ray diffraction data and the Canadian Light source for access. The infrastructure at the Centre for Blood Research is supported by the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund. J.N.K. is a recipient of a Michael Smith Foundation for Health Research Scholar award. P.R. was supported by a Leopoldina-Postdoc-Fellowship from the Deutsche Akademie der Naturforscher Leopoldina. Financial support from the Canadian Institutes for Health Research (CIHR grant MOP-136940) is gratefully acknowledged by J.N.K. and S.G.W. H.M. is supported by a Canadian Blood Services-MITACS fellowship and an NSERC CREATE Nanomat scholarship.
Author information
Authors and Affiliations
Contributions
P.R. performed the majority of the screening and characterization work as well as preparing figures and tables and providing a first draft of the manuscript; L.S. performed all work related to three-dimensional structural analysis; H.M. performed the majority of the testing with RBCs, in conjunction with I.C., and contributed to the writing; C.M.-L. performed much of the bioinformatics analysis; S.J.H. provided expertise in metagenomic analysis and supervision of the bioinformatics; J.N.K. supervised all work related to testing with RBCs and edited the manuscript; S.G.W. conceived the project, supervised the enzymology and coordinated the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The University of British Columbia has filed a patent related to this work on which P.R., J.N.K. and S.G.W. are named as authors.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion, Supplementary References, Supplementary Figs. 1–22 and Supplementary Tables 1–18.
Supplementary Datasets 1 and 2
FpGalNAcDeAc_D2ext glycan array data and circular dichroism spectrometer data.
Rights and permissions
About this article
Cite this article
Rahfeld, P., Sim, L., Moon, H. et al. An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nat Microbiol 4, 1475–1485 (2019). https://doi.org/10.1038/s41564-019-0469-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0469-7
This article is cited by
-
Enzymatic conversion of human blood group A kidneys to universal blood group O
Nature Communications (2024)
-
Host genetic regulation of human gut microbial structural variation
Nature (2024)
-
Turning universal O into rare Bombay type blood
Nature Communications (2023)
-
Replacing renal function using bioengineered tissues
Nature Reviews Bioengineering (2023)
-
Genome sequence and Carbohydrate Active Enzymes (CAZymes) repertoire of the thermophilic Caldicoprobacter algeriensis TH7C1T
Microbial Cell Factories (2022)