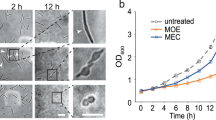
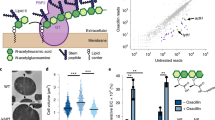
Abstract
Peptidoglycan (PGN) is the major component of the bacterial cell wall, a structure that is essential for the physical integrity and shape of the cell. Bacteria maintain cell shape by directing PGN incorporation to distinct regions of the cell, namely, through the localization of late-stage PGN synthesis proteins. These include two key protein families, SEDS transglycosylases and bPBP transpeptidases, proposed to function in cognate pairs. Rod-shaped bacteria have two SEDS–bPBP pairs, involved in elongation and division. Here, we elucidate why coccoid bacteria, such as Staphylococcus aureus, also possess two SEDS–bPBP pairs. We determined that S. aureus RodA–PBP3 and FtsW–PBP1 probably constitute cognate pairs of interacting proteins. A lack of RodA–PBP3 resulted in more spherical cells due to deficient sidewall PGN synthesis, whereas depletion of FtsW–PBP1 arrested normal septal PGN incorporation. Although PBP1 is an essential protein, a mutant lacking PBP1 transpeptidase activity is viable, showing that this protein has a second function. We propose that the FtsW–PBP1 pair has a role in stabilizing the divisome at midcell. In the absence of these proteins, the divisome appears as multiple rings or arcs that drive lateral PGN incorporation, leading to cell elongation. We conclude that RodA–PBP3 and FtsW–PBP1 mediate sidewall and septal PGN incorporation, respectively, and that their activity must be balanced to maintain coccoid morphology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author on request.
Code availability
The in-house developed image analysis software is available in the GitHub repository: https://github.com/BacterialCellBiologyLab/PyFRET.
References
Egan, A. J., Cleverley, R. M., Peters, K., Lewis, R. J. & Vollmer, W. Regulation of bacterial cell wall growth. FEBS J. 284, 851–867 (2017).
Chastanet, A. & Carballido-Lopez, R. The actin-like MreB proteins in Bacillus subtilis: a new turn. Front. Biosci. (Schol. Ed.) 4, 1582–1606 (2012).
Domínguez-Escobar, J. et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011).
Garner, E. C. et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011).
van Teeffelen, S. et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl Acad. Sci. USA 108, 15822–15827 (2011).
den Blaauwen, T., Hamoen, L. W. & Levin, P. A. The divisome at 25: the road ahead. Curr. Opin. Microbiol. 36, 85–94 (2017).
Egan, A. J. & Vollmer, W. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277, 8–28 (2013).
Massidda, O., Novakova, L. & Vollmer, W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ. Microbiol. 15, 3133–3157 (2013).
Pinho, M. G., Kjos, M. & Veening, J.-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 11, 601–614 (2013).
Perez, A. J. et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA 116, 3211–3220 (2019).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 (2011).
Taguchi, A. et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019).
Emami, K. et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol. 2, 16253 (2017).
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016).
Scheffers, D. J. & Pinho, M. G. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69, 585–607 (2005).
Monteiro, J. M. et al. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055 (2015).
Pereira, S. F., Henriques, A. O., Pinho, M. G., de Lencastre, H. & Tomasz, A. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189, 3525–3531 (2007).
Pinho, M. G., de Lencastre, H. & Tomasz, A. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 182, 1074–1079 (2000).
Pereira, S. F. F., Henriques, A. O., Pinho, M. G., de Lencastre, H. & Tomasz, A. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol. Microbiol. 72, 895–904 (2009).
Sassine, J. et al. Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 106, 304–318 (2017).
Monteiro, J. M. et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005).
Kuru, E. et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl. 51, 12519–12523 (2012).
Levin, P. A., Kurtser, I. G. & Grossman, A. D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl Acad. Sci. USA 96, 9642–9647 (1999).
Jorge, A. M., Hoiczyk, E., Gomes, J. P. & Pinho, M. G. EzrA contributes to the regulation of cell size in Staphylococcus aureus. PLoS ONE 6, e27542 (2011).
den Blaauwen, T., de Pedro, M. A., Nguyen-Disteche, M. & Ayala, J. A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32, 321–344 (2008).
Aaron, M. et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64, 938–952 (2007).
Rohs, P. D. A. et al. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726 (2018).
Cho, H., Uehara, T. & Bernhardt, T. G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014).
Du, S., Pichoff, S. & Lutkenhaus, J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl Acad. Sci. USA 113, E5052–E5061 (2016).
Modell, J. W., Kambara, T. K., Perchuk, B. S. & Laub, M. T. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, e1001977 (2014).
Lund, V. A. et al. Molecular coordination of Staphylococcus aureus cell division. eLife 7, e32057 (2018).
Veiga, H. & Pinho, M. G. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl. Environ. Microbiol. 75, 3034–3038 (2009).
Oshida, T. & Tomasz, A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174, 4952–4959 (1992).
Pereira, P., Veiga, H., Jorge, A. & Pinho, M. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl. Environ. Microbiol. 76, 4346–4353 (2010).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual 2nd edn (Cold Spring Harbor Laboratory Press, 1989).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Filipe, S. R., Tomasz, A. & Ligoxygakis, P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 6, 327–333 (2005).
Reed, P. et al. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog. 11, e1004891 (2015).
Kuru, E., Tekkam, S., Hall, E., Brun, Y. V. & Van Nieuwenhze, M. S. Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 10, 33–52 (2015).
Pereira, A. R. et al. FtsZ-dependent elongation of a coccoid bacterium. mBio 7, e00908-16 (2016).
Chen, H., Puhl, H. L. 3rd, Koushik, S. V., Vogel, S. S. & Ikeda, S. R. Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys. J. 91, L39–L41 (2006).
Hoppe, A., Christensen, K. & Swanson, J. A. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 83, 3652–3664 (2002).
Acknowledgements
We thank L. Moreira for hosting laboratory work at the Instituto Superior Técnico, University of Lisbon, Lisbon, Portugal, S. R. Filipe (FCT-NOVA) and R. Carballido-Lopez (INRA) for helpful discussions, S. Bonucci and E. M. Tranfield (Electron Microscopy Facility, IGC) for technical expertise and sample processing, and A. Bernardo, I. Jorge, D. Kądziołka and K. Witana for help in the construction of some plasmids. This study was funded by the European Research Council through grant ERC-2017-CoG-771709 (to M.G.P.), Project LISBOA-01-0145FEDER-007660 Microbiologia Molecular, Estrutural e Celular (to ITQB-NOVA), the Portuguese Platform of Bioimaging PPBI-POCI-01-0145-FEDER-022122, researcher contract no. IF/00386/2015 (to F.F.) and FCT fellowships SFRH/BPD/95031/2013 (to N.T.R.) and SFRH/BD/52204/2013 (to A.C.T.). Whole-genome sequencing analysis at the Genomics Unit of Instituto Gulbenkian de Ciencia was supported by the ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI—‘Fundos Europeus Estruturais e de Investimento’ from ‘Programa Operacional Regional Lisboa 2020’—and by national funds from FCT—‘Fundação para a Ciência e a Tecnologia’.
Author information
Authors and Affiliations
Contributions
N.T.R., A.C.T. and M.G.P. designed the research. N.T.R. and A.C.T. performed all of the experiments, with the exception of the FLIM data acquisition and analysis, which was performed by F.F., and the HPLC muropeptide analysis performed by A.J. B.M.S. developed software for image analysis of the seFRET data. N.T.R., A.C.T. and P.R. constructed the strains. A.J. and P.R. analysed the whole-genome sequencing data. J.M.M. and A.R.P. performed the preliminary experiments and analysed the data. R.G.S. performed the preliminary experiments. M.S.V.N. contributed new reagents (FDAAs). N.T.R., A.C.T. and M.G.P. analysed the overall data. B.M.S. analysed the seFRET data quantified by in-house developed software. N.T.R., A.C.T. and M.G.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11, Supplementary Tables 1–3, Supplementary Video legends and Supplementary References.
Supplementary Video 1
ColFtsWi time-lapse imaging.
Supplementary Video 2
ColPBP1i time-lapse imaging.
Supplementary Video 3
ColPBP1TP time-lapse imaging.
Supplementary Video 4
ColPBP1TP time-lapse imaging.
Rights and permissions
About this article
Cite this article
Reichmann, N.T., Tavares, A.C., Saraiva, B.M. et al. SEDS–bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat Microbiol 4, 1368–1377 (2019). https://doi.org/10.1038/s41564-019-0437-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0437-2