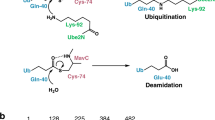
Abstract
The bacterial pathogen Legionella pneumophila modulates host immunity using effectors translocated by its Dot/Icm transporter to facilitate its intracellular replication. A number of these effectors employ diverse mechanisms to interfere with protein ubiquitination, a post-translational modification essential for immunity. Here, we have found that L. pneumophila induces monoubiquitination of the E2 enzyme UBE2N by its Dot/Icm substrate MavC(Lpg2147). We demonstrate that MavC is a transglutaminase that catalyses covalent linkage of ubiquitin to Lys92 and Lys94 of UBE2N via Gln40. Similar to canonical transglutaminases, MavC possess deamidase activity that targets ubiquitin at Gln40. We identified Cys74 as the catalytic residue for both ubiquitination and deamidation activities. Furthermore, ubiquitination of UBE2N by MavC abolishes its activity in the formation of K63-type polyubiquitin chains, which dampens NF-κB signalling in the initial phase of bacterial infection. Our results reveal an unprecedented mechanism of modulating host immunity by modifying a key ubiquitination enzyme by ubiquitin transglutamination.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the conclusions of this study are included in this published article along with its Supplementary Information files, and are also available from the corresponding author upon request.
References
Isberg, R. R., O’Connor, T. J. & Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009).
Losick, V. P. & Isberg, R. R. NF-κB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203, 2177–2189 (2006).
Bartfeld, S. et al. Temporal resolution of two-tracked NF-κB activation by Legionella pneumophila. Cell Microbiol. 11, 1638–1651 (2009).
Abu-Zant, A. et al. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 9, 246–264 (2007).
Ge, J. et al. A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc. Natl Acad. Sci. USA 106, 13725–13730 (2009).
Losick, V. P., Haenssler, E., Moy, M. Y. & Isberg, R. R. LnaB: a Legionella pneumophila activator of NF-κB. Cell Microbiol. 12, 1083–1097 (2010).
Jiang, X. & Chen, Z. J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12, 35–48 (2011).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Maculins, T., Fiskin, E., Bhogaraju, S. & Dikic, I. Bacteria–host relationship: ubiquitin ligases as weapons of invasion. Cell Res. 26, 499–510 (2016).
Cui, J. et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218 (2010).
Qiu, J. & Luo, Z. Q. Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Microbiol. 15, 591–605 (2017).
Qiu, J. et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 (2016).
Bhogaraju, S. et al. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649 (2016).
Kotewicz, K. M. et al. A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell. Host Microbe 21, 169–181 (2017).
Hodge, C. D., Spyracopoulos, L. & Glover, J. N. Ubc13: the Lys63 ubiquitin chain building machine. Oncotarget 7, 64471–64504 (2016).
O’Connor, T. J., Adepoju, Y., Boyd, D. & Isberg, R. R. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl Acad. Sci. USA 108, 14733–14740 (2011).
Huang, L. et al. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 13, 227–245 (2010).
Eddins, M. J., Carlile, C. M., Gomez, K. M., Pickart, C. M. & Wolberger, C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 (2006).
Lorand, L. & Graham, R. M. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 (2003).
Keillor, J. W., Clouthier, C. M., Apperley, K. Y., Akbar, A. & Mulani, A. Acyl transfer mechanisms of tissue transglutaminase. Bioorg. Chem. 57, 186–197 (2014).
Klock, C. & Khosla, C. Regulation of the activities of the mammalian transglutaminase family of enzymes. Protein Sci. 21, 1781–1791 (2012).
Eckert, R. L. et al. Transglutaminase regulation of cell function. Physiol. Rev. 94, 383–417 (2014).
Chen, Z. J. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 (2005).
Zhou, H. et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427, 167–171 (2004).
Liu, X., Fitzgerald, K., Kurt-Jones, E., Finberg, R. & Knipe, D. M. Herpesvirus tegument protein activates NF-κB signaling through the TRAF6 adaptor protein. Proc. Natl Acad. Sci. USA 105, 11335–11339 (2008).
Sanada, T. et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 (2012).
Mukherjee, S. et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214 (2006).
Stewart, M. D., Ritterhoff, T., Klevit, R. E. & Brzovic, P. S. E2 enzymes: more than just middle men. Cell Res. 26, 423–440 (2016).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 (2009).
Zou, W. et al. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem. Biophys. Res. Commun. 336, 61–68 (2005).
Takeuchi, T. & Yokosawa, H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem. Biophys. Res. Commun. 336, 9–13 (2005).
Machida, Y. J. et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 (2006).
Rape, M. & Kirschner, M. W. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 432, 588–595 (2004).
Ravid, T. & Hochstrasser, M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat. Cell Biol. 9, 422–427 (2007).
Valleau, D. et al. Discovery of ubiquitin deamidases in the pathogenic arsenal of Legionella pneumophila. Cell Rep. 23, 568–583 (2018).
Urbanus, M. L. et al. Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12, 893 (2016).
Berger, K. H. & Isberg, R. R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 (1993).
Luo, Z. Q. & Isberg, R. R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl Acad. Sci. USA 101, 841–846 (2004).
Xu, L. et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 6, e1000822 (2010).
Sheedlo, M. J. et al. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc. Natl Acad. Sci. USA 112, 15090–15095 (2015).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Gotze, M. et al. StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass. Spectrom. 23, 76–87 (2012).
Li, X. et al. Negative regulation of hepatic inflammation by the soluble resistance-related calcium-binding protein via signal transducer and activator of transcription 3. Front. Immunol. 8, 709 (2017).
Acknowledgements
The authors thank R. Isberg (Tufts School of Medicine) for bacterial strains, F. Shao (National Institute for Biological Sciences, Beijing) for plasmids, and R. Moore and A. Shukla at the Pacific Northwest National Laboratory (PNNL) for technical support. This work was supported by National Institutes of Health grants R01AI127465(ZQL), R21AI117205 (PJH and ZQL) and GM103493. Part of this work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a Department of Energy (DOE) office of Biological and Environmental Research (BER) National User Facility located at the PNNL.
Author information
Authors and Affiliations
Contributions
N.G. and Z.-Q.L. conceived the ideas for this work. N.G. planned and performed experiments. N.G., E.S.N., P.J.H. and Z.-Q.L. interpreted the data. E.S.N. performed mass spectrometric analyses. N.G. and Z.-Q.L. wrote the manuscript and all authors provided editorial input.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11.
Supplementary Table 1
Proteins potentially modified by 3×Ub-AA in mammalian cells.
Rights and permissions
About this article
Cite this article
Gan, N., Nakayasu, E.S., Hollenbeck, P.J. et al. Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N. Nat Microbiol 4, 134–143 (2019). https://doi.org/10.1038/s41564-018-0282-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0282-8
This article is cited by
-
An expanded lexicon for the ubiquitin code
Nature Reviews Molecular Cell Biology (2023)
-
Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC
Nature Communications (2020)
-
Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors
Nature Communications (2020)
-
Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination
Nature Communications (2020)
-
Bacterial virulence mediated by orthogonal post-translational modification
Nature Chemical Biology (2020)