Abstract
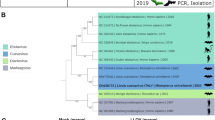
Here we describe the complete genome of a new ebolavirus, Bombali virus (BOMV) detected in free-tailed bats in Sierra Leone (little free-tailed (Chaerephon pumilus) and Angolan free-tailed (Mops condylurus)). The bats were found roosting inside houses, indicating the potential for human transmission. We show that the viral glycoprotein can mediate entry into human cells. However, further studies are required to investigate whether exposure has actually occurred or if BOMV is pathogenic in humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
08 November 2018
In the version of this Article originally published, the bat species for 12 individuals were incorrectly identified in Supplementary Table 1 and 2. After resequencing the MT-CytB and MT-CO1 segments and reviewing the data, the authors have corrected the errors for these 12 animals. In the amended version of the Supplementary Information, Supplementary Tables 1 and 2 have been replaced to include the corrected host species information. None of the 12 bats affected were positive for the Bombali virus, and the conclusions of the study are therefore unchanged.
References
Burk, R. et al. Neglected filoviruses. FEMS Microbiol. Rev. 40, 494–519 (2016).
Marí Saéz, A. et al. Investigating the zoonotic origin of the West African Ebola epidemic.EMBO Mol. Med. 7, 17–23 (2015).
Lo, T. Q., Marston, B. J., Dahl, B. A. & De Cock, K. M. Ebola: anatomy of an epidemic. Annu. Rev. Med. 68, 359–370 (2017).
Leroy, E. M. et al. Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576 (2005).
Pourrut, X. et al. Spatial and temporal patterns of Zaire ebolavirus antibody prevalence in the possible reservoir bat species. J. Infect. Dis. 196, S176–S183 (2007).
Hayman, D. T. et al. Ebola virus antibodies in fruit bats, Ghana, West Africa. Emerg. Infect. Dis. 18, 1207–1209 (2012).
Yuan, J. et al. Serological evidence of ebolavirus infection in bats, China. Virol. J. 9, 236 (2012).
Negredo., A. et al. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog. 7, e1002304 (2011).
Jayme, S. I. et al. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol. J. 12, 107 (2015).
Bào, Y. et al. Implementation of objective PASC-derived taxon demarcation criteria for official classification of filoviruses. Viruses 9, 106 (2017).
Swanepoel, R. et al. Experimental inoculation of plants and animals with Ebola virus. Emerg. Infect. Dis. 2, 321–325 (1996).
Leendertz, S. A. J. Testing new hypotheses regarding ebolavirus reservoirs. Viruses 8, 30 (2016).
Carette, J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011).
Côté, M. et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348 (2011).
Miller, E. H. et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31, 1947–1960 (2012).
Ng, M. et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife 4, e11785 (2015).
Bornholdt, Z. A. et al. Host-primed Ebola virus GP exposes a hydrophobic NPC1 receptor-binding pocket, revealing a target for broadly neutralizing antibodies. mBio 7, e02154-15 (2016).
Wang, H. et al. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1.Cell 164, 258–268 (2016).
Pappalardo, M. et al. Conserved differences in protein sequence determine the human pathogenicity of Ebolaviruses. Sci. Rep. 6, 23743 (2016).
Miranda, M. E. & Miranda, N. L. Reston ebolavirus in humans and animals in the Philippines: a review. J. Infect. Dis. 204, S757–S760 (2011).
Bale, S. et al. Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. J. Virol. 87, 10385–10388 (2013).
Reid, S. P. et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80, 5156–5167 (2006).
Volchkov, V. E., Blinov, V. M. & Netesov, S. V. The envelope glycoprotein of Ebola virus contains an immunosuppressive-like domain similar to oncogenic retroviruses. FEBS Lett. 305, 181–184 (1992).
Schoepp, R. J., Rossi, C. A., Khan, S. H., Goba, A. & Fair, J. N. Undiagnosed acute viral febrile illnesses, Sierra Leone. Emerg. Infect. Dis. 20, 1176–1182 (2014).
Towner, J. S. et al. Marburg virus infection detected in a common African bat. PLoS ONE 2, e764 (2007).
Yang, X. L. et al. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg. Infect. Dis. 23, 482–486 (2017).
Amman, B. R. et al. Marburgvirus resurgence in Kitaka Mine bat population after extermination attempts, Uganda.Emerg. Infect. Dis. 20, 1761–1764 (2014).
Townzen, J. S., Brower, A. V., & Judd, D. D. Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences.Med. Vet. Entomol. 22, 386–393 (2008).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Towner, J. S. et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome.J. Virol. 78, 4330–4341 (2004).
Jääskeläinen, A. J. et al. Development and evaluation of a real-time EBOV-L-RT-qPCR for detection of Zaire ebolavirus.J. Clin. Virol. 67, 56–58 (2015).
Anthony, S. J. et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8, e00373-17 (2017).
Briese, T. et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio 6, e01491-15 (2015).
Spence, J. S., Krause, T. B., Mittler, E., Jangra, R. K. & Chandran, K. Direct visualization of Ebola virus fusion triggering in the endocytic pathway. mBio 7, e01857-15 (2016).
Wong, A. C., Sandesara, R. G., Mulherkar, N., Whelan, S. P. & Chandran, K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84, 163–175 (2010).
Whelan, S. P. J., Ball, L. A., Barr, J. N. & Wertz, G. T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl Acad. Sci. USA 92, 8388–8392 (1995).
Kleinfelter, L. M. et al. Haploid genetic screen reveals a profound and direct dependence on cholesterol for hantavirus membrane fusion. mBio 6, e00801 (2015).
Chandran, K., Sullivan, N. J., Felbor, U., Whelan, S. P. & Cunningham, J. M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005).
Ng, M. et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann-Pick C1.Virology 468–470, 637–646 (2014).
Petrey, D. et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling.Proteins 53, S430–S435 (2003).
Li, W., Jaroszewski, L. & Godzik, A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283 (2001).
King, D. P. et al. Humoral immune responses to phocine herpesvirus-1 in Pacific harbor seals (Phoca vitulina richardsii) during an outbreak of clinical disease.Vet. Microbiol. 80, 1–8 (2001).
Acknowledgements
We thank the government of Sierra Leone for permission to conduct this work; the Sierra Leone district and community stakeholders for their support and for allowing us to perform sampling in their districts and communities; the Bombali Ministry of Health and Sanitation and Ministry of Agriculture district officers, field teams and regional lead including M. LeBreton, F. Jean Louis, K. Kargbo, L.A.M. Kenny, V. Lungay, W. Robert, E. Amara, D. Kargbo, V. Merewhether-Thompson, M. Kanu, E. Lavallie, A. Bangura, M. Turay, F.V. Bairoh, M. Sinnah and S. Yonda for performing sample collection; Yongai Saah Bona for administrative and logistic support; laboratory staff for assistance with processing the samples, including M. Coomber and O. Kanu (University of Makeni) and V. Ontiveros (UC Davis); T. O’Rourke, D. O’ Rourke (Metabiota) and D. Greig (UC Davis) for assistance with data entry, B. Lee for bioinformatics assistance and J. Morrison and A. Rasmussen for technical guidance (Columbia University); N. Randhawa for map graphics (UC Davis); and W. Karesh and J. Epstein (EcoHealth Alliance) for global input into study design. This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number GHN-A-OO-09-00010-00) and by support from the National Institutes of Health (GM030518, S10OD012351, S10OD021764 and GM109018-05).
Author information
Authors and Affiliations
Contributions
T.G. helped design the study, collected, analysed and interpreted the data, helped with the literature search and with writing the manuscript. S.J.A. helped design the study, collected, analysed and interpreted the data and helped with writing the manuscript. A.G. supervised the in-country activities, including obtaining permissions, and helped with sample and data collection. B.H.B. helped design the study, analysed the data and helped with the literature search. J.B. performed and supervised all aspects of field activities and helped with sample and data collection. A.T-B. and M.N.B. collected and analysed data. H.W. collected, analysed and interpreted data and helped with writing the manuscript. J.K.D. collected the data. E.L. and M.G. collected, analysed and interpreted data and helped with writing the manuscript. R.K.J. collected and analysed the data and helped with writing the manuscript. V.A.D. collected and analysed the data. G.L. collected, analysed and interpreted the data and helped with writing the manuscript. B.R.S. collected the data. A.J., B.O.K., S.K. and W.B. provided project permissions and logistical support. C.M. helped design the study and collected the data. S.S. analysed and interpreted the data and helped with writing the manuscript. C.K-J. helped design the study. K.S. oversaw the project. E.M.R. oversaw the project and analysed the data. K.C. collected and analysed the data and helped with writing the manuscript. W.I.L. analysed and interpreted the data and helped with writing the manuscript. J.A.K.M. oversaw and designed the project and helped with writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Supplementary Figures 1–6.
Rights and permissions
About this article
Cite this article
Goldstein, T., Anthony, S.J., Gbakima, A. et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3, 1084–1089 (2018). https://doi.org/10.1038/s41564-018-0227-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0227-2
This article is cited by
-
Selective replication and vertical transmission of Ebola virus in experimentally infected Angolan free-tailed bats
Nature Communications (2024)
-
Kenyan Free-Tailed Bats Demonstrate Seasonal Birth Pulse Asynchrony with Implications for Virus Maintenance
EcoHealth (2024)
-
Isolation and genome characterization of Lloviu virus from Italian Schreibers’s bats
Scientific Reports (2023)
-
A random priming amplification method for whole genome sequencing of SARS-CoV-2 virus
BMC Genomics (2022)
-
Glycan shield of the ebolavirus envelope glycoprotein GP
Communications Biology (2022)