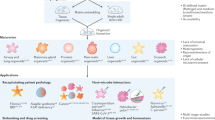
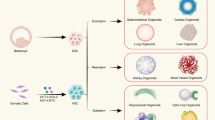
Abstract
In recent years considerable progress has been made in the development of faithful procedures for the differentiation of human pluripotent stem cells (hPSCs). An important step in this direction has also been the derivation of organoids. This technology generally relies on traditional three-dimensional culture techniques that exploit cell-autonomous self-organization responses of hPSCs with minimal control over the external inputs supplied to the system. The convergence of stem cell biology and bioengineering offers the possibility to provide these stimuli in a controlled fashion, resulting in the development of naturally inspired approaches to overcome major limitations of this nascent technology. Based on the current developments, we emphasize the achievements and ongoing challenges of bringing together hPSC organoid differentiation, bioengineering and ethics. This Review underlines the need for providing engineering solutions to gain control of self-organization and functionality of hPSC-derived organoids. We expect that this knowledge will guide the community to generate higher-grade hPSC-derived organoids for further applications in developmental biology, drug screening, disease modelling and personalized medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rudnick, D. Regulation and localization in the hind limb bud of the chick embryo. Anat. Rec. 94, 492 (1946).
Saunders, J. W. An experimental study of the distribution, orientation, and tract specificity of feather germs in the wing of the chick embryo. Anat. Rec. 99, 647 (1947).
Moscona, A. & Moscona, H. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J. Anat. 86, 287–301 (1952).
Weiss P, T. A. Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc. Natl Acad. Sci. USA 46, 1177–1185 (1960).
Auerbach, R. & Grobstein, C. Inductive interaction of embryonic tissues after dissociation and reaggregation. Exp. Cell Res. 15, 384–397 (1958).
Simian, M. & Bissell, M. J. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 216, 31–40 (2017).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Xia, Y. & Izpisua Belmonte, J. C. Design approaches for generating organ constructs. Cell Stem Cell 24, 877–894 (2019).
Thomson, Ja. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008).
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Pasca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Sakaguchi, H. et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 6, 8896 (2015).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–110 (2011).
Dye, B. R. et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, e05098 (2015).
Koehler, K. R. et al. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 35, 583–589 (2017).
Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Armstrong, P. B. Cell sorting out: the self-assembly of tissues in vitro. Crit. Rev. Biochem. Mol. Biol. 24, 119–149 (1989).
van Den Brink, S. C. et al. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Develeopment 141, 4231–4242 (2014).
Ten Berge, D. et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3, 508–518 (2008).
van den Brink, S. C. et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582, 405–409 (2020).
Dessaud, E., McMahon, A. P. & Briscoe, J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489–2503 (2008).
Meinhardt, A. et al. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports 3, 987–999 (2014).
Renner, M. et al. Self‐organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329 (2017).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Rakic, P. Extrinsic cytological determinants of basket and stellate cell dendritic pattern in the cerebellar molecular layer. J. Comp. Neurol. 146, 335–354 (1972).
Volpato, V. et al. Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-derived neurons: a multi-site omics study. Stem Cell Reports 11, 897–911 (2018).
Phipson, B. et al. Evaluation of variability in human kidney organoids. Nat. Methods 16, 79–87 (2019).
Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D. & Brivanlou, A. H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854 (2014).
Martyn, I., Kanno, T. Y., Ruzo, A., Siggia, E. D. & Brivanlou, A. H. Self-organization of a human organizer by combined Wnt and nodal signaling. Nature 558, 132–135 (2018).
Ma, Z. et al. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 6, 7413 (2015).
Kim, H. Y. & Nelson, C. M. Extracellular matrix and cytoskeletal dynamics during branching morphogenesis. Organogenesis 8, 56–64 (2012).
Vianello, S. & Lutolf, M. P. Understanding the mechanobiology of early mammalian development through bioengineered models. Dev. Cell 48, 751–763 (2019).
Cruz-Acuña, R. et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326–1335 (2017).
Garreta, E. et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 18, 397–405 (2019).
Lancaster, M. A. et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666 (2017).
Storm, C., Pastore, J. J., MacKintosh, F. C., Lubensky, T. C. & Janmey, P. A. Nonlinear elasticity in biological gels. Nature 435, 191–194 (2005).
Uzel, S. G. M. et al. Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small 12, 612–622 (2016).
Manfrin, A. et al. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods 16, 640–648 (2019).
Cederquist, G. Y. et al. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 37, 436–444 (2019).
DeForest, C. A. & Anseth, K. S. Photoreversible patterning of biomolecules within click-based hydrogels. Angew. Chemie Int. Ed. 51, 1816–1819 (2012).
Guvendiren, M. & Burdick, J. A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 3, 792 (2012).
Vincent, L. G., Choi, Y. S., Alonso-Latorre, B., Del Álamo, J. C. & Engler, A. J. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol. J. 8, 472–484 (2013).
Xia, B. & Yanai, I. A periodic table of cell types. Development 146, dev169854 (2019).
Roost, M. S. et al. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Reports 4, 1112–1124 (2015).
Nowotschin, S. et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Mayr, U., Serra, D. & Liberali, P. Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development 146, dev176727 (2019).
Hannezo, E. & Heisenberg, C. P. Mechanochemical feedback loops in development and disease. Cell 178, 12–25 (2019).
Bailles, A. et al. Genetic induction and mechanochemical propagation of a morphogenetic wave. Nature 572, 467–473 (2019).
Alt, S., Ganguly, P. & Salbreux, G. Vertex models: from cell mechanics to tissue morphogenesis. Philos. Trans. R. Soc. B 372, 20150520 (2017).
Latorre, E. et al. Active superelasticity in three-dimensional epithelia of controlled shape. Nature 563, 203–208 (2018).
Okuda, S. et al. Strain-triggered mechanical feedback in self-organizing optic-cup morphogenesis. Sci. Adv. 4, eaau1354 (2018).
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H. & Reiner, O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515–522 (2018).
Bershteyn, M. et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20, 435–449 (2017).
Forbes, T. A. et al. Patient-iPSC-derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am. J. Hum. Genet. 102, 816–831 (2018).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Bian, S. et al. Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15, 631–639 (2018).
Ogawa, J., Pao, G. M., Shokhirev, M. N. & Verma, I. M. Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220–1229 (2018).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Przepiorski, A. et al. A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 11, 470–484 (2018).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Sabbagh, M. F. et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. eLife 7, e36187 (2018).
Takebe, T. et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 16, 556–565 (2015).
Czerniecki, S. M. et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940 (2018).
Taguchi, A. & Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746 (2017).
Mansour, A. A. et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441 (2018).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Takebe, T. et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 21, 2661–2670 (2017).
Van den Berg, C. W. et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 10, 751–765 (2018).
Campisi, M. et al. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 180, 117–129 (2018).
Shirure, V. S. et al. Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab Chip 18, 3687–3702 (2018).
Song, J., Miermont, A., Lim, C. T. & Kamm, R. D. A 3D microvascular network model to study the impact of hypoxia on the extravasation potential of breast cell lines. Sci. Rep. 8, 17949 (2018).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019).
Mark, A. et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 5, eaaw2459 (2019).
Noor, N. et al. 3D personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 6, 1900344 (2019).
Workman, M. J. et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49–59 (2017).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Koike, H. et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary. Nature 574, 112–116 (2019).
Esch, M. B., Mahler, G. J., Stokol, T. & Shuler, M. L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 14, 3081–3092 (2014).
Bauer, S. et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 7, 14620 (2017).
Osaki, T., Uzel, S. G. M. & Kamm, R. D. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 4, eaat5847 (2018).
Achberger, K. et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. eLife 8, e46188 (2019).
Tao, T. et al. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip 19, 948–958 (2019).
Workman, M. J. et al. Enhanced utilization of induced pluripotent stem cell–derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 5, 669–677 (2018).
Lee, K. K. et al. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip 18, 3079–3085 (2018).
Wang, Y. et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 18, 3606–3616 (2018).
Bredenoord, A. L., Clevers, H. & Knoblich, J. A. Human tissues in a dish: the research and ethical implications of organoid technology. Science 355, eaaf9414 (2017).
Munsie, M., Hyun, I. & Sugarman, J. Ethical issues in human organoid and gastruloid research. Development 144, 942–945 (2017).
Hyun, I. Engineering ethics and self-organizing models of human development: opportunities and challenges. Cell Stem Cell 21, 718–720 (2017).
Van de Poel, I. & van Gorp, A. C. The need for ethical reflection in engineering design. Sci. Technol. Hum. Values 31, 333–360 (2006).
Sample, M. et al. Multi-cellular engineered living systems: building a community around responsible research on emergence. Biofabrication 11, 043001 (2019).
Chuva de Sousa Lopes, S. M. Accelerating maturation of kidney organoids. Nat. Mater. 18, 303–304 (2019).
Miura, Y. & Pașca, S. P. Polarizing brain organoids. Nat. Biotechnol. 37, 377–378 (2019).
Wilson, H. V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 5, 245–258 (1907).
Harrison, R. G. Observations on the living developing nerve fiber. Exp. Biol. Med. 4, 140–143 (1906).
Strangeways, T. S. P. & Fell, H. B. Experimental studies on the differentiation of embryonic tissues growing in vivo and in vitro.—II. The development of the isolated early embryonic eye of the fowl when cultivated in vitro. Proc. R. Soc. B Biol. Sci. 100, 273–283 (1926).
Trowell, O. A. A modified technique for organ culture in vitro. Exp. Cell Res. 6, 246–248 (1954).
Streuli, C. H. & Bissell, M. J. Expression of extracellular matrix components is regulated by substratum. J. Cell Biol. 110, 1405–1415 (1990).
Roca-Cusachs, P., Conte, V. & Trepat, X. Quantifying forces in cell biology. Nat. Cell Biol. 19, 742–751 (2017).
Grashoff, C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010).
Legant, W. R. et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 7, 969–971 (2010).
Campàs, O. et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183–189 (2014).
Kumar, S. et al. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J. 90, 3762–3773 (2006).
Etournay, R. et al. TissueMiner: a multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. eLife 5, e14334 (2016).
Acknowledgements
We thank SOLIDCAM ESTUDIO for support with figure illustrations. This work has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (StG-2014-640525_REGMAMKID to N.M., CoG-2013-616480_TensionControl to X.T., CoG-2016-725722_OVOGROWTH to S.M.C.S.L. and StG-757710 to M.A.L.). This research has been supported by the EFSD/Boehringer Ingelheim European Research Programme in Microvascular Complications of Diabetes, and EIT Health under grant ID 20366 (R2U‐Tox‐Assay) to E.G. and N.M. R.D.K. received support from the US National Science Foundation (CBET-0939511). M.A.L. received funding from the Medical Research Council (MC_UP_1201/9). R.W. received support from the NIH (R01EB024591 and R01EB025256) and the NSF (0939511 and 1446474) research grants. X.T. is also supported by the European Commission (project H2020-FETPROACT-01-2016-731957). I.H. is funded by the Greenwall Foundation’s ‘Making a Difference’ grant. This work also received funding from the Spanish Ministry of Economy and Competitiveness (MINECO)/FEDER (SAF2015-72617-EXP to N.M. and SAF2017-89782-R to N.M. and PGC2018-099645-B-I00 to X.T.), Generalitat de Catalunya and CERCA programme (2017 SGR 1306 to N.M. and SGR-2017-01602 to X.T.), Asociación Española contra el Cáncer (AECC) (LABAE16006 to N.M.). N.M. is also supported by Instituto de Salud Carlos III (Tercel, Cardiocel and ACE2ORG). The Institute for Bioengineering of Catalonia is the recipient of a ‘Centro de Excelencia Severo Ochoa’ award from the MINECO (funded by the Agencia Estatal de Investigación: SEV2014-0425 and CEX2018-000789-S) and MIT-SPAIN "la Caixa” Foundation SEED FUND project “Bioenginering Against Cancer” funded by MISTI Global Seed Funds and “la Caixa” Foundation.
Author information
Authors and Affiliations
Contributions
N.M. conceived the outline of the Review, and wrote and revised the manuscript. E.G. wrote and revised the manuscript. R.D.K. contributed in the section on engineering vascularization and revised the manuscript. S.M.C.S.L. contributed on the section of single-cell transcriptomics and commented on the manuscript. M.A.L. contributed to the section of understanding self-organization and symmetry breaking. R.W. commented on the manuscript. X.T. contributed to the section of probing mechanics in hPSC-organoids. I.H. wrote the section of engineering ethics and commented on the manuscript. N.M. and E.G. created the figures.
Corresponding author
Ethics declarations
Competing interests
R.D.K. is co-founder and has a substantial financial interest in AIM Biotech, and receives research support from Amgen, Biogen and Gore.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Garreta, E., Kamm, R.D., Chuva de Sousa Lopes, S.M. et al. Rethinking organoid technology through bioengineering. Nat. Mater. 20, 145–155 (2021). https://doi.org/10.1038/s41563-020-00804-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-020-00804-4
This article is cited by
-
Patient-derived organoids in human cancer: a platform for fundamental research and precision medicine
Molecular Biomedicine (2024)
-
Microfluidic high-throughput 3D cell culture
Nature Reviews Bioengineering (2024)
-
Vascularised cardiac spheroids-on-a-chip for testing the toxicity of therapeutics
Scientific Reports (2024)
-
A microfluidic platform integrating functional vascularized organoids-on-chip
Nature Communications (2024)
-
Innovative explorations: unveiling the potential of organoids for investigating environmental pollutant exposure
Environmental Science and Pollution Research (2024)