Abstract
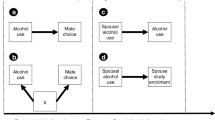
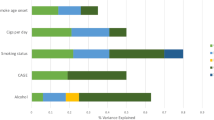
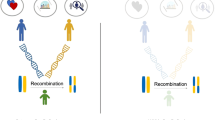
To investigate assortative mating (AM), participation bias and socioeconomic status (SES) with respect to the genetics of behavioural and psychiatric traits, we estimated AM signatures using gametic phase disequilibrium and within-spouses and within-siblings polygenic risk score correlation analyses, also performing a SES conditional analysis. The cross-method meta-analysis identified AM genetic signatures for multiple alcohol-related phenotypes, bipolar disorder, major depressive disorder, schizophrenia and Tourette syndrome. Here, after SES conditioning, we observed changes in the AM genetic signatures for maximum habitual alcohol intake, frequency of drinking alcohol and Tourette syndrome. We also observed significant gametic phase disequilibrium differences between UK Biobank mental health questionnaire responders versus non-responders for major depressive disorder and alcohol use disorder. These results highlight the impact of AM, participation bias and SES on the polygenic risk of behavioural and psychiatric traits, particularly in alcohol-related traits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All results used to make conclusions discussed in this study are provided as Supplementary Material. All GWAS data are publicly available on their respective websites: UK Biobank (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access); Psychiatric Genomics Consortium (https://pgc.unc.edu/for-researchers/download-results/); Million Veteran Program (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v8.p1); Pan-UK Biobank (https://pan.ukbb.broadinstitute.org/).
Code availability
The majority of the analyses were conducted using previously developed tools. These were described in Methods with their corresponding references. Custom R scripts developed for the simulation analysis and for estimating expected cross-method results are available on Zenodo (https://doi.org/10.5281/zenodo.10476703).
References
Wendt, F. R. et al. Characterizing the effect of background selection on the polygenicity of brain-related traits. Genomics 113, 111–119 (2021).
Plomin, R. & von Stumm, S. Polygenic scores: prediction versus explanation. Mol. Psychiatry 27, 49–52 (2022).
Yengo, L. et al. Imprint of assortative mating on the human genome. Nat. Hum. Behav. 2, 948–954 (2018).
Taylor, A. E. et al. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J. Epidemiol. 47, 1207–1216 (2018).
Nordsletten, A. E. et al. Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry 73, 354–361 (2016).
Peyrot, W. J., Robinson, M. R., Penninx, B. W. & Wray, N. R. Exploring boundaries for the genetic consequences of assortative mating for psychiatric traits. JAMA Psychiatry 73, 1189–1195 (2016).
Border, R. et al. Cross-trait assortative mating is widespread and inflates genetic correlation estimates. Science 378, 754–761 (2022).
Lamers, F. et al. Sociodemographic and psychiatric determinants of attrition in the Netherlands Study of Depression and Anxiety (NESDA). Compr. Psychiatry 53, 63–70 (2012).
Gorman, E. et al. Assessing the representativeness of population-sampled health surveys through linkage to administrative data on alcohol-related outcomes. Am. J. Epidemiol. 180, 941–948 (2014).
Tyrrell, J. et al. Genetic predictors of participation in optional components of UK Biobank. Nat. Commun. 12, 886 (2021).
Pirastu, N. et al. Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. 53, 663–671 (2021).
Widiger, T. A. Personality and psychopathology. World Psychiatry 10, 103–106 (2011).
Gelernter, J. & Polimanti, R. Genetics of substance use disorders in the era of big data. Nat. Rev. Genet. 22, 712–729 (2021).
Lichter, D. T. and Qian Z. in The Study of Assortative Mating: Theory, Data, and Analysis. Analytical Family Demography (ed. Schoen, R.) (Springer International Publishing, 2019).
Russell, G. et al. Selection bias on intellectual ability in autism research: a cross-sectional review and meta-analysis. Mol. Autism 10, 9 (2019).
Sareen, J., Afifi, T. O., McMillan, K. A. & Asmundson, G. J. Relationship between household income and mental disorders: findings from a population-based longitudinal study. Arch. Gen. Psychiatry 68, 419–427 (2011).
Shah, N. et al. National or population level interventions addressing the social determinants of mental health—an umbrella review. BMC Public Health 21, 2118 (2021).
Hill, W. D. et al. Genome-wide analysis identifies molecular systems and 149 genetic loci associated with income. Nat. Commun. 10, 5741 (2019).
Wendt, F. R. et al. Multivariate genome-wide analysis of education, socioeconomic status and brain phenome. Nat. Hum. Behav. 5, 482–496 (2021).
Sullivan, P. F. et al. Psychiatric genomics: an update and an agenda. Am. J. Psychiatry 175, 15–27 (2018).
Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Nievergelt, C. M. et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 10, 4558 (2019).
Howe, L. J. et al. Assortative mating and within-spouse pair comparisons. PLoS Genet. 17, e1009883 (2021).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Aschard, H., Vilhjalmsson, B. J., Joshi, A. D., Price, A. L. & Kraft, P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet 96, 329–339 (2015).
Deak, J. D. et al. Genome-wide investigation of maximum habitual alcohol intake in US veterans in relation to alcohol consumption traits and alcohol use disorder. JAMA Netw. Open 5, e2238880 (2022).
Polimanti, R. et al. Evidence of causal effect of major depression on alcohol dependence: findings from the psychiatric genomics consortium. Psychol. Med. 49, 1218–1226 (2019).
Agrawal, A. et al. Assortative mating for cigarette smoking and for alcohol consumption in female Australian twins and their spouses. Behav. Genet 36, 553–566 (2006).
Grant, J. D. et al. Spousal concordance for alcohol dependence: evidence for assortative mating or spousal interaction effects? Alcohol Clin. Exp. Res 31, 717–728 (2007).
Reynolds, C. A., Barlow, T. & Pedersen, N. L. Alcohol, tobacco and caffeine use: spouse similarity processes. Behav. Genet 36, 201–215 (2006).
Rhule-Louie, D. M. & McMahon, R. J. Problem behavior and romantic relationships: assortative mating, behavior contagion, and desistance. Clin. Child Fam. Psychol. Rev. 10, 53–100 (2007).
Colbert, S. M. C. et al. Declining autozygosity over time: an exploration in over 1 million individuals from three diverse cohorts. Am. J. Hum. Genet 110, 1008–1014 (2023).
Bulmer, M. G. The effect of selection on genetic variability: a simulation study. Genet. Res. 28, 101–117 (1976).
Wendt, F. R. et al. Using phenotype risk scores to enhance gene discovery for generalized anxiety disorder and posttraumatic stress disorder. Mol. Psychiatry 27, 2206–2215 (2022).
Davis, K. A. S. et al. Mental health in UK Biobank—development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open 6, e18 (2020).
Hugh-Jones, D., Verweij, K. J. H., St. Pourcain, B. & Abdellaoui, A. Assortative mating on educational attainment leads to genetic spousal resemblance for polygenic scores. Intelligence 59, 103–108 (2016).
Levey, D. F. et al. Reproducible genetic risk loci for anxiety: results from approximately 200,000 participants in the million veteran program. Am. J. Psychiatry 177, 223–232 (2020).
Levey, D. F. et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 24, 954–963 (2021).
Wendt, F. R. et al. Modeling the longitudinal changes of ancestry diversity in the Million Veteran Program. Hum. Genomics 17, 46 (2023).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Tsai, J. & Rosenheck, R. A. Risk factors for homelessness among US veterans. Epidemiol. Rev. 37, 177–195 (2015).
Collins, S. E. Associations between socioeconomic factors and alcohol outcomes. Alcohol Res. 38, 83–94 (2016).
Okbay, A. et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 54, 437–449 (2022).
Robinson, M. R. et al. Genetic evidence of assortative mating in humans. Nat. Hum. Behav. 1, 0016 (2017).
Wang, X. et al. Polygenic risk prediction: why and when out-of-sample prediction R2 can exceed SNP-based heritability. Am. J. Hum. Genet 110, 1207–1215 (2023).
Conomos, M. P., Reiner, A. P., Weir, B. S. & Thornton, T. A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet 98, 127–148 (2016).
1000 Genomes Project Consortiumet al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Li, J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Duncan, L. et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858 (2017).
Otowa, T. et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol. Psychiatry 21, 1485 (2016).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Lam, M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019).
Forstner, A. J. et al. Genome-wide association study of panic disorder reveals genetic overlap with neuroticism and depression. Mol. Psychiatry 26, 4179–4190 (2021).
Duncan, L. E. et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry 23, 666–673 (2018).
Yu, D. et al. Interrogating the genetic determinants of Tourette’s syndrome and other tic disorders through genome-wide association studies. Am. J. Psychiatry 176, 217–227 (2019).
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF–GC) & OCD Collaborative Genetics Association (OCDCGA) Studies. Revealing the complex genetic architecture of obsessive–compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Psychiatric Genomics Consortium (PCG) Genome-wide association statistics https://pgc.unc.edu/for-researchers/download-results/ (2023).
Kranzler, H. R. et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10, 1499 (2019).
Gelernter, J. et al. Genome-wide association study of maximum habitual alcohol intake in >140,000 U.S. European and African American veterans yields novel risk loci. Biol. Psychiatry 86, 365–376 (2019).
Zhou, H. et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry 77, 1072–1080 (2020).
Stein, M. B. et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat. Genet. 53, 174–184 (2021).
Veterans Administration (VA) Million Veteran Program (MVP) summary results from omics studies. dbGaP https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v8.p1 (2022).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience 8, giz082 (2019).
International HapMap Consortiumet al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Wu, Y. et al. Estimating genetic nurture with summary statistics of multigenerational genome-wide association studies. Proc. Natl Acad. Sci. USA 118, e2023184118 (2021).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48 (2010).
Cabrera-Mendoza, B., Yengo, L. & Polimanti, R. Custom R scripts for ‘The impact of assortative mating, participation bias, and socioeconomic status on the polygenic risk of behavioral and psychiatric traits’. Zenodo https://doi.org/10.5281/zenodo.10476703 (2024).
Townsend, P., Phillimore, P. & Beattie, A. Health and Deprivation: Inequality and the North 1st edn (Croom Helm, 1988).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Acknowledgements
We thank the participants and the investigators involved in the UK Biobank, Million Veteran Program and the Psychiatric Genomics Consortium for making their data publicly available. This research was conducted using the UK Biobank Resource (application reference number 58146). The authors acknowledge support from the National Institutes of Health (R21 DC018098, R33 DA047527 and RF1 MH132337 to R.P. and K99 AG078503 to G.A.P.), One Mind (Rising Star Award to R.P.) and the American Foundation for Suicide Prevention (PDF-1-022-21 to B.C.-M.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
B.C.-M., L.Y. and R.P. designed this study. B.C.-M. conducted the analysis. B.C.-M. and R.P. wrote the paper. F.R.W., G.A.P. and L.Y. critically revised the paper. All authors contributed to interpretation of the data. L.Y. and R.P. contributed equally to this work and jointly supervised the study.
Corresponding authors
Ethics declarations
Competing interests
R.P. is paid for their editorial work on the journal Complex Psychiatry and reports a research grant from Alkermes. The other authors declare no competing interests.
Peer review
Peer review information
Nature Human Behaviour thanks Lea Davis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods and Fig. 1.
Supplementary Table
Supplementary Tables 1–21.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cabrera-Mendoza, B., Wendt, F.R., Pathak, G.A. et al. The impact of assortative mating, participation bias and socioeconomic status on the polygenic risk of behavioural and psychiatric traits. Nat Hum Behav (2024). https://doi.org/10.1038/s41562-024-01828-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41562-024-01828-5