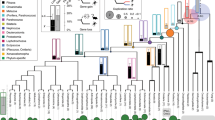
Abstract
The animal kingdom shows an astonishing diversity, the product of over 550 million years of animal evolution. The current wealth of genome sequence data offers an opportunity to better understand the genomic basis of this diversity. Here we analyse a sampling of 102 whole genomes including >2.6 million protein sequences. We infer major genomic patterns associated with the variety of animal forms from the superphylum to phylum level. We show that a remarkable amount of gene loss occurred during the evolution of two major groups of bilaterian animals, Ecdysozoa and Deuterostomia, and further loss in several deuterostome lineages. Deuterostomes and protostomes also show large genome novelties. At the phylum level, flatworms, nematodes and tardigrades show the largest reduction of gene complement, alongside gene novelty. These findings paint a picture of evolution in the animal kingdom in which reductive evolution at the protein-coding level played a major role in shaping genome composition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Publicly available genomes are listed in Supplementary Table 1.
Code availability
All the scripts used in this study can be found at https://github.com/CristiGuijarro/ComparativeGenomics.
Change history
27 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41559-020-1159-9
References
Egger, B. et al. A transcriptomic-phylogenomic analysis of the evolutionary relationships of flatworms. Curr. Biol. 25, 1347–1353 (2015).
Jékely, G., Paps, J. & Nielsen, C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. EvoDevo 6, 1 (2015).
Marlétaz, F., Peijnenburg, K. T. C. A., Goto, T., Satoh, N. & Rokhsar, D. S. A new spiralian phylogeny places the enigmatic arrow worms among gnathiferans. Curr. Biol. 29, 312–318.e3 (2019).
Giribet, G. New animal phylogeny: future challenges for animal phylogeny in the age of phylogenomics. Org. Divers. Evol. 16, 419–426 (2016).
Halanych, K. M. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 35, 229–256 (2004).
Richter, D. J., Fozouni, P., Eisen, M. B. & King, N. Gene family innovation, conservation and loss on the animal stem lineage. eLife 7, e34226 (2018).
Paps, J. What makes an animal? The molecular quest for the origin of the animal kingdom. Integr. Comp. Biol. 58, 654–665 (2018).
Grau-Bové, X. et al. Dynamics of genomic innovation in the unicellular ancestry of animals. eLife 6, e26036 (2017).
Lankester, E. R. Degeneration. A Chapter in Darwinism (Macmillan and Company, 1880).
Denoeud, F. et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–1385 (2010).
Tsai, I. J. et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496, 57–63 (2013).
Zmasek, C. M. & Godzik, A. This déjà vu feeling—analysis of multidomain protein evolution in eukaryotic genomes. PLoS Comput. Biol. 8, e1002701 (2012).
Moore, A. D. & Bornberg-Bauer, E. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 29, 787–796 (2012).
Albalat, R. & Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016).
Paps, J. & Holland, P. W. H. Nat. Commun. 9, 1730 (2018).
Dunwell, T. L., Paps, J. & Holland, P. W. H. Novel and divergent genes in the evolution of placental mammals. Proc. R. Soc. B 284, 20171357 (2017).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, 61–65 (2007).
Arakawa, K. No evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc. Natl Acad. Sci. USA 113, E3057 (2016).
Yoshida, Y. et al. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol. 15, e2002266 (2017).
The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017).
Simakov, O. et al. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531 (2013).
Luo, Y.-J. et al. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat. Ecol. Evol. 2, 141–151 (2018).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Laumer, C. E. et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol. 25, 2000–2006 (2015).
Kocot, K. M. On 20 years of Lophotrochozoa. Org. Divers. Evol. 16, 329–343 (2016).
Kocot, K. M. et al. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256–282 (2017).
Acknowledgements
We thank I. Maeso for comments on the manuscript. C.G.-C. and J.P. received funding from the School of Biological Sciences (University of Essex).
Author information
Authors and Affiliations
Contributions
C.G.-C., J.P. and P.W.H.H. designed the study and analyses. C.G.-C. performed the analyses. All the authors wrote the manuscript. C.G.-C. drew additional animal outlines (the flatworm and the rotifer, both of which are Public Domain Dedication 1.0 license and No Copyright, see Supplementary Information) in Fig. 1.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and Tables 1–3.
Supplementary Data 1
HG assignment for each genome, and how many proteins each genome has assigned to each cluster for core novel, core lost, novel and lost HGs for each phylum. Files are named by clade position in the phylogeny followed by the assigned clade name.
Supplementary Data 2
List of protein sequence headers or IDs assigned to each novel and lost HG for each phylum.
Supplementary Data 3
GO and model protein assignment for each of the core HGs analysed in the pipeline. There is a file for each of the core lost and core novel HGs for each clade analysed. GOs were mined from InterProScan.
Supplementary Data 4
GO and model protein assignment for each of the HGs analysed in the pipeline. There is a file for each of the lost and novel HGs for each clade analysed.
Supplementary Data 5
BLAST checks of the novel HGs (all the proteins in each HG) for each phylum with the RefSeq database. Thresholds include 1 × 10−6 e-value and >50% identity matches.
Supplementary Data 6
Available as an xlsx sorted document. The number of protein class Panther hits and other GOs for each node. The numbers between nodes cannot be compared due to the varying model organisms used and protein class annotations; however, each node should be compared internally within the same model organisms. Model organism for protein class used is the same as written in Supplementary Data 3 and 4. Orphan HGs were excluded from the analysis. Urochordate, cephalochordate, hemichordate, platyhelminth and tardigrade novelties were also excluded from these data due to the lack of protein classes in the Panther analysis for specific model organisms. GOs and protein IDs are available in Supplementary Data 3 and 4.
Rights and permissions
About this article
Cite this article
Guijarro-Clarke, C., Holland, P.W.H. & Paps, J. Widespread patterns of gene loss in the evolution of the animal kingdom. Nat Ecol Evol 4, 519–523 (2020). https://doi.org/10.1038/s41559-020-1129-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-020-1129-2
This article is cited by
-
ContScout: sensitive detection and removal of contamination from annotated genomes
Nature Communications (2024)
-
Macroevolutionary dynamics of gene family gain and loss along multicellular eukaryotic lineages
Nature Communications (2024)
-
Hagfish genome elucidates vertebrate whole-genome duplication events and their evolutionary consequences
Nature Ecology & Evolution (2024)
-
Gene loss during a transition to multicellularity
Scientific Reports (2023)
-
Genomes of fungi and relatives reveal delayed loss of ancestral gene families and evolution of key fungal traits
Nature Ecology & Evolution (2023)