Abstract
Males and females often differ in their fitness optima for shared traits that have a shared genetic basis, leading to sexual conflict. Morphologically differentiated sex chromosomes can resolve this conflict and protect sexually antagonistic variation, but they accumulate deleterious mutations. However, how sexual conflict is resolved in species that lack differentiated sex chromosomes is largely unknown. Here we present a chromosome-anchored genome assembly for rainbow trout (Oncorhynchus mykiss) and characterize a 55-Mb double-inversion supergene that mediates sex-specific migratory tendency through sex-dependent dominance reversal, an alternative mechanism for resolving sexual conflict. The double inversion contains key photosensory, circadian rhythm, adiposity and sex-related genes and displays a latitudinal frequency cline, indicating environmentally dependent selection. Our results show sex-dependent dominance reversal across a large autosomal supergene, a mechanism for sexual conflict resolution capable of protecting sexually antagonistic variation while avoiding the homozygous lethality and deleterious mutations associated with typical heteromorphic sex chromosomes.
Similar content being viewed by others
Main
Differential selection on male and female individuals results in sexual antagonism, with profound implications for genome evolution, adaptation and the maintenance of fitness variation1,2,3,4. In the classical model of sex chromosome evolution, sexually antagonistic polymorphisms accumulate in linkage disequilibrium with the sex-determining locus, driving selection for reduced recombination and differentiation of the sex chromosomes (X/Y, or Z/W), and the eventual degradation of the hemizygous chromosome through accumulation of deleterious mutations5,6. While this model explains sexual conflict resolution in some species, many taxa lack morphologically differentiated sex chromosomes5; how sexually antagonistic variation is maintained in these species is largely unknown4,7. Additionally, recent theoretical work has predicted that sexual conflict may be better resolved by autosomal variation8, a prediction supported by genome-wide mapping of sexually antagonistic polymorphisms3. Thus, mechanisms that maintain sexual conflict polymorphisms on the autosomes must be common, yet the only known such mechanism—sex-dependent dominance reversal7,9, where the favourable allele in each sex is dominant in that sex—has been observed only for a single maturity gene in Atlantic salmon4 and for genome-wide variation for fitness in seed beetles10. As a result, the mechanisms maintaining sexually antagonistic variation in the absence of differentiated sex chromosomes and their consequence for adaptive and genome evolution are unresolved.
Autosomal inversion supergenes controlling alternative reproductive tactics and resembling sex chromosomes have been identified in some taxa, but typically suffer from homozygous lethality11,12 and concomitant chromosomal degradation13,14. Balancing selection can facilitate the evolution of new dominance patterns and these, along with epistasis, are important features of inversion polymorphisms15,16. Typical of taxa with homomorphic sex chromosomes5,8,17, salmonids have undergone frequent sex chromosome turnover18, and sex-reversed males (XY females) have been suggested to occur19,20, both of which are predicted to limit divergence of the sex chromosomes5,21 and their ability to accumulate and protect sexual conflict polymorphisms8,17.
Oncorhynchus mykiss (rainbow trout) is a salmonid fish species that expresses two contrasting life-history strategies: resident rainbow trout live entirely in freshwater, while anadromous steelhead trout migrate to the ocean to mature, returning to freshwater to reproduce. The decision to either mature early or delay maturation and migrate is a complex heritable trait, requiring the integration of internal (energy status/adiposity) and external (photoperiod and temperature) signals22. Survival of migratory juveniles to adulthood is very low, but fecundity of anadromous females can exceed that of resident females by an order of magnitude22. In contrast, males can mature early as freshwater residents, avoiding the high mortality associated with marine migration and employing a sneaker mating strategy to access paternity22. This sex-specific trade-off between reproduction and survival results in a greater frequency of anadromy in females23 and may drive sexual conflict over alternative migratory tactics with a shared genetic basis. Previous work has identified a major effect locus on chromosome Omy05 that associates with migratory phenotypes24,25, but the structure and composition of this region as well as its ability to resolve sexual conflict are unknown. Here we generate a chromosome-level genome assembly for rainbow trout and use it to characterize a large autosomal inversion supergene on Omy05, and show that it mediates sexual conflict over migratory tendency via sex-dependent dominance reversal. Lacking homozygote lethality, this inversion complex provides a mechanism for maintaining polygenic sexually antagonistic variation while avoiding the deleterious mutation load accumulated by differentiated sex chromosomes.
Results and discussion
Rainbow trout genome reveals structural rearrangements
To characterize the genetic and genomic architecture of complex traits, it is important to construct a high-quality, chromosome-anchored whole-genome sequence; this is a challenge in rainbow trout owing to the salmonid-specific whole-genome duplication event (Ss4R) approximately 80–125 million years ago (Ma)26,27,28, and subsequent expansion of repetitive elements27. We created an assembly (GCA_002163495.1) containing 139,800 scaffolds with an N50 of 1.67 Mb and a total of 2.18 gigabases (Gb), representing more than 90% of the predicted length (2.4 Gb) of the rainbow trout genome29. High-density linkage analysis, based on the mapping of 46,174 single-nucleotide polymorphisms (SNPs) in a pedigree of 146 full-sib families with 5,716 individuals, together with long-range data from the Dovetail Genomics Chicago library sequencing30, was used to order and orient 7,868 scaffolds within linkage groups, producing 29 chromosome-length sequences containing 1.92 Gb (88.5%) of the genome assembly (Supplementary Information, Section 1; Supplementary Tables 1 and 2), representing a substantial improvement over the previous assembly26 (Supplementary Table 3). Most scaffolds not anchored to chromosomes (in total 131,938 scaffolds spanning 229 Mb of sequence) consisted of repetitive sequences. Overall, repetitive sequences account for 57.1% of the rainbow trout genome (Supplementary Information, Section 5; Extended Data Fig. 1), similar to the 59.9% previously reported for Atlantic salmon27. Annotation by the National Center for Biotechnology Information (NCBI) RefSeq pipeline predicted 53,383 genes, of which 42,884 are protein-coding (NCBI Oncorhynchus mykiss, Annotation Release 100). Analysis of homeologous regions resulting from the salmon-specific duplication revealed 88 collinear blocks along 29 chromosomes (Fig. 1 and Supplementary Information, Section 2).
a–d, Inner to outer circles: a, Homeologous regions in the rainbow trout genome subdivided into 88 collinear blocks along 29 chromosomes. The red rectangles represent blocks of sequences without identifiable duplicated regions elsewhere in the genome. b, Genomic similarity (in 1-Mb intervals) between duplicated regions. c, Frequency of Tc1-mariner transposon elements in the rainbow trout genome. b,c, Red, high; yellow, medium; green, low sequence similarity or frequency, respectively. d, High-resolution female (red) and male (blue) linkage maps constructed from the analysis of 44,910 markers genotyped in a family material of 5,716 fish.
Linkage mapping detailed striking recombination differences between the sexes across the genome (Fig. 1d and Supplementary Information, Section 3), as documented previously31, resolved variable chromosome numbers associated with centric fusions or fissions in rainbow trout (Extended Data Fig. 2) and revealed recombination patterns suggestive of large polymorphic inversions on chromosomes Omy05 and Omy20 (Fig. 2 and Extended Data Fig. 3). Haplotypes tagging these rearrangements were identified and used to classify the parents of the mapping families. Subsequent linkage mapping in homozygous parent families disclosed the structure of the inversions, while linkage mapping in families from heterozygous parents documented almost complete repression of recombination across the rearrangements (Extended Data Fig. 3). The Omy05 rearrangement is characterized by two adjacent inversions of 21.99 and 32.83 Mb, of which the first is pericentric, reversing the centromere. The alternative karyotypes in the Omy05 double inversion were categorized as ancestral (A) or rearranged (R) based on their sequence and structural synteny relative to the Atlantic salmon, coho salmon and Arctic char genomes, and linkage maps for Chinook, chum and sockeye salmon (Extended Data Fig. 4), as well as their prior associations with the anadromous (A) and resident (R) life histories24,25. Because the draft genome assembly was made from a homozygous RR individual, we generated a second genome assembly for a male homozygous for the ancestral karyotype using long-read nanopore sequencing to determine more exact breakpoints for the Omy05 rearrangements (Extended Data Fig. 5 and Supplementary Information, Section 4). The Omy20 inversion region is smaller, approximately 14.0 Mb (Extended Data Fig. 3), and has not been described previously or associated with any known phenotype.
a, Divergence (FST, black dots, window 1,500 bp) between individuals homozygous for alterative Omy05 karyotypes, RR and AA, reveals a large area of elevated divergence that coincides with two large linkage map discontinuities (red lines). These indicate two large inversions, the first of which is pericentromeric, meaning it moves the centromere (the black circle at base of the plot). Key candidate genes with related functions are spread across the two inversions; the colours depict trait-relevant functions. b, Sequence divergence (dXY) between karyotypes, 50-kb windows sliding by 15 kb. c,d, Sequence diversity (π) among AA (blue) and RR (red) individuals, across 1.5-kb windows; the dotted line is the genome-wide mean π. e, Map of western North America showing all 83 sampling locations, with the bars showing the relative frequencies of the A (blue) and R (red) Omy05 rearrangement karyotypes. The purple numbers indicate locations where whole-genome resequenced individuals were selected. Credit: map insets: global location and location of Big Creek, Monterey County, California101. f, Frequency of inversion karyotype R as a function of latitude among a subset of 42 populations of North American rainbow trout with migratory access to the ocean. Point sizes are proportional to sample size (n) with the bars showing ± s.e.m. Weighted least squares regression line, y = 0.04x – 1.21, adjusted R2 = 0.51.
Whole-genome resequencing of 31 individuals homozygous for the Omy05 inversion sampled across the species’ native range revealed decreased diversity within (π) and increased relative (FST) and absolute (dXY) divergence between the 9 AA and 22 RR karyotype individuals (Fig. 2a–d, Supplementary Information, Section 7, and Supplementary Table 4c). A sharp rise in divergence occurred at the inversion boundaries coincident with the breakpoints in the linkage map, with maximal divergence in the pericentromeric region peaking in a 2.5-Mb region of inversion 1 (Fig. 2a). This pericentromeric region also displays a pronounced decrease in sequence diversity (Fig. 2c,d) and is enriched for genes containing segregating missense mutations (pericentromeric region, 47%; rest of rearrangement, 30.5%). Dating of coding sequence (CDS) divergence across the inversions suggested that they have been maintained for approximately 1.5 million years (Extended Data Fig. 6; see Methods). There was no evidence for different ages of the two inversions, leaving the order of occurrence unresolved. Furthermore, no evidence of differences in the dating estimates was found between the centre and inversion breakpoints either from the dating of coding regions or FST, consistent with the double inversion forming a very strong barrier to recombination.
Sex-dependent dominance of a migration supergene and sexual conflict resolution
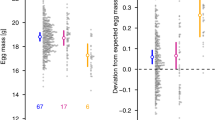
Sex-specific migratory optima are expected to result in intra-locus sexual conflict in rainbow trout where there is a shared genetic basis to migratory tendency. Previous studies have associated genetic markers on Omy05 with migratory traits in rainbow trout24,25; thus, the double inversion could potentially have sex-specific effects on life history. Therefore, we tested the role inversion karyotype plays in mediating sexual conflict and whether sex-dependent dominance contributes to its resolution. Mark–recapture analysis of > 2,600 individually tagged O. mykiss from a small stream (Big Creek, California, USA; Fig. 2e) showed that sex and Omy05 karyotype (AA, AR or RR) both strongly influence the probability of an individual migrating to the ocean (Fig. 3). These results are consistent with shifts in sex and Omy05 karyotype frequencies within the stream among trout across the migratory size range (>100 mm; Fig. 3b,c). The best-fit model of migratory tendency includes sex-dependent dominance of the Omy05 rearrangement, where the karyotype with the highest predicted fitness for each sex is dominant in that sex (delta Akaike’s Information Criterion (ΔAIC) = 4.90; Supplementary Information, Section 8 and Supplementary Table 5). At peak size for juvenile marine migration (approximately 150 mm fork length), homozygous AA females were more than twice as likely to be detected emigrating as RR females (70.9 versus 26.7%), with a reduced difference in males (45.3 versus 31.8). In the full genotype model, heterozygous females were estimated to emigrate at the same rate as homozygous AA females (complete dominance, d/a = 0.97; Fig. 3a) while heterozygous males more closely resembled RR males (partial dominance, d/a = 0.48; Supplementary Table 5 and Extended Data Fig. 7). Such asymmetric sex-dependent dominance reversal is predicted when the strength of antagonistic selection differs between the sexes9, and has also been observed in age at maturity in Atlantic salmon4.
a, Migration of female (left) and male (right) juveniles with AA (blue squares), AR (orange triangles) and RR (red circles) genotypes tagged in freshwater at peak size for individual migration, as estimated from detections at the in-stream fixed antennas, with generalized additive model fits (95% confidence intervals). b, Proportions of Omy05 rearrangement genotypes. c, Proportions of genetic sex among all females (solid lines) and males (dashed lines) sampled in Big Creek and ordered by size. b,c, Individuals were binned into length categories of approximately 170 individuals to calculate the mean values, with the bars showing ± s.e.m.
The observed sex-dependent dominance reversal of the Omy05 double inversion and its role in resolving sexual conflict over migratory tendency could reflect the homeology between the centromeric half of the sex chromosome (Omy29: 0–26.28 Mb) and Omy05, including 6.45 Mb of inversion 1 (Fig. 1 and Extended Data Figs. 4 and 8). However, it is unknown what role, if any, the sex chromosomes of rainbow trout play in sexual conflict resolution. The sex-determining locus, sdY32, of salmonids including rainbow trout, is located within a transposon cassette, leading to frequent chromosome turnover and a lack of homology among sex chromosomes throughout the family and within Atlantic salmon20,33. Theory predicts sex chromosome translocations to be driven by the need to resolve sexual conflict or avoid mutation load accumulation34,35, but the frequent turnover of salmonid sex chromosomes may limit their capacity to protect sexual conflict polymorphisms relative to typical heteromorphic sex chromosomes5. Because even homomorphic sex chromosomes may be differentiated at the molecular level36, we next tested for signatures of divergence and the accumulation of sexually antagonistic variation in the rainbow trout sex chromosome, Omy29, to determine if homeology with the sex chromosome could explain the apparent role played by the Omy05 supergene in sexual conflict resolution.
Elevated genetic differentiation (FST) between the sexes is predicted around sexually antagonistic loci in linkage disequilibrium with the sex determiner37. High between-sex FST among 38 wild individuals at Omy29: 5 Mb (Fig. 4a), as well as bacterial artificial chromosome sequence alignment, supported this region as the location of the sdY transposon cassette38 (Supplementary Information, Section 6), placing sdY within the region of homeology to Omy05, but outside the rearrangement. It is noteworthy that this region is highly rearranged, creating a barrier to homeologue recombination with Omy29 (Extended Data Figs. 4a,c and 8b). Linkage analysis confirmed that male recombination is strongly localized towards the telomere on Omy29 (Fig. 4d and Extended Data Fig. 8a), as has been observed previously in rainbow trout39, Atlantic salmon27 and other taxa40,41. Thus, if XY divergence is limited only by recombination in males, divergence should be seen across most of the Y chromosome. In contrast, divergence was largely restricted to the region between sdY and the centromere, where recombination is also low in females (Fig. 4a,d). These results suggest that XY differentiation is further limited by recombination in sex-reversed males (XY females), supporting the fountain of youth hypothesis for ‘ever young’ Y chromosomes21 and maintaining a large pseudoautosomal region with limited potential to resolve sexual conflict.
a–c, Between-sex divergence along the sex chromosome, Omy29, shown by FST (a), coverage in 500-bp windows (b) and exon coverage (c) is concentrated in the 5-Mb region between the centromere (0 Mb) and the male determiner, sdY. Red lines denote the 0.1% highest genome-wide M:F Fst (a) and P = 0.05 with Bonferroni correction (b,c). d, This region has low recombination in both males and females. e, Relative to the autosomes, genes with maximal expression in oocyte or testis are not enriched on the Y chromosome (number of genes ± s.e.m., where expression in gonadal tissue was compared to expression across 13 other tissues and TPM > 5). P values from one-tailed Fisher’s exact test.
To test for enrichment of genes on Omy29 with sex-specific effects, we defined male and female benefit genes as those with maximum expression in testis and oocyte, respectively, compared to a panel of 13 somatic tissues. The Y chromosome was not enriched for male benefit genes (178 out of 919 Y genes compared to 9,005 out of 47,415 autosomal genes, −log2 transcripts per million (TPM), one-tailed Fisher’s exact test, P = 0.513; Fig. 4e), consistent with XY recombination and/or frequent turnover preventing enrichment of male benefit genes. Conversely, low recombination between X and Y with weak sexual antagonism can lead to feminization of the Y chromosome8. However, there was also no enrichment in female benefit genes (173 out of 919 Y genes compared to 8,091 out of 47,415 autosomal genes, −log2 TPM, Fisher’s exact test, P = 0.29), with the same proportion of both testis and oocyte genes as expected if they were evenly distributed across the chromosomes (18–19%; Fig. 4e). These results were further supported by the similar distributions of sequence coverage between males and females, consistent with the Y chromosome not having gained or retained genes not present on the X chromosome (Fig. 4b,c). The symmetrical levels of gene loss between the Y chromosome and its autosomal orthologue in coho salmon, Oki29 (ref. 42; 15 and 12%, respectively), also indicate no excess of gene loss on the rainbow trout Y chromosome. Finally, we found little evidence of structural rearrangements on Omy29 that are predicted to accompany Y chromosome recombination shutdown43. Instead, Omy29 chromosome structure is highly conserved with the distantly related Atlantic salmon and char (divergence times approximately 20 Ma; Extended Data Fig. 8c,d), whereas the comparative regions in all other mapped Pacific salmon species are rearranged (Extended Data Fig. 8e,f).
Together, the absence of XY differentiation, sex-biased gene enrichment, rearrangements or other signals predicted by the classical model of sex chromosome evolution5,6 suggest that selection against deleterious mutation load, rather than the resolution of sexual conflict17,21, has driven Y chromosome evolution in rainbow trout. In contrast, the absence of homozygote lethality of either karyotype of the Omy05 inversion allows within-karyotype recombination to purge deleterious mutations, thus avoiding degradation. Thus, sex-dependent dominance reversal of the Omy05 rearrangement provides an alternative autosomal mechanism to resolve sexual conflict while avoiding the mutation load associated with the canonical heteromorphic sex chromosome system.
Gene composition of the Omy05 rearrangement
Inversion supergenes allow coadapted variants to avoid being broken up by recombination44. The 55-Mb double inversion on Omy05 contains 1,091 protein-coding genes; a single causative gene or mutation is probably not responsible for the observed phenotypic association. Genes in the inversion include key adiposity, circadian rhythm/entrainment, photosensory and age at maturity genes associated with migratory behaviour and seasonal timing of maturation (Fig. 2a and Supplementary Table 6a–c; see Methods). Visual-related pathways are known to be overrepresented among genes differentially expressed between resident and anadromous rainbow trout45, reflecting the importance of light in the timing of smoltification and maturation. The inversion includes the master regulator of circadian rhythm, CLOCK, and the visual pigment, OPN4, which is expressed in the saccus vasculosus, the organ that controls photoperiodism and so reproductive timing in fish46. Further, the highly divergent pericentromeric region contains a cluster of photosensory and circadian genes, PDCL, PPEF2, RX3 and MAPK10 (JNK3)47,48,49 (Fig. 2a) and a serotonin receptor, 5-HT2B, which affects both behaviour and retinal development50,51. The homeologous regions of CLOCK and MAPK10 on Omy01 and Omy12, respectively, have previously been implicated in migratory phenotypes in northern populations52,53, supporting a role for these genes in trait divergence and suggesting that selection on gene duplicates may contribute to a more diffuse architecture in the north, independent of the Omy05 inversion.
The mechanism causing sex-dependent dominance reversal across the inversion is unknown; however, the Omy05 inversion contains candidate genes with sex-specific effects and/or with sex-biased expression in teleosts. Notably, DMRTA2 is a duplicate of the sexual dimorphism gene Doublesex54, which is expressed in the developing gonads and pituitary55,56 with greater adult expression in the testes than ovaries of teleost fish57,58. In zebrafish, DMRTA2 regulates terminal differentiation of corticotropes and gonadotropes, through which it may accelerate gonadal development and so influence maturation timing55. DMRTA2 regulates cells expressing pro-opiomelanocortin (pomc55), which is differentially expressed between migratory forms of rainbow trout45 and Atlantic salmon59. DMRTA2 and pomc are also differentially expressed in rx3 mutant zebrafish60, resulting in disrupted circadian rhythm48. Two other genes known to have strong sex-specific effects and to affect maturation are found in the inversion AMH, a gene involved in the differentiation of rainbow trout gonads61,62, and NR5A2, which is involved in oestrogen biosynthesis and inhibition of adipogenesis through its regulation of CYP19a1 (refs. 63,64). This is important because adiposity is a key determiner of migratory behaviour that is strongly and consistently associated with divergent migratory and maturation phenotypes in salmonid fish4,22,65,66. Along with NR5A2, the Omy05 rearrangement contains the adiposity genes RORC1, RXRA and LEPR67,68,69 (Fig. 2a), all of which are associated with age at maturity in humans70. In addition, CENPR (ITGB3BP, NRIF3), a coactivator of oestrogen receptor alpha71 and enhancer of nuclear receptors, thyroid hormones and retinoid X receptors (RXR72), is also found near the distal breakpoint of inversion 2. Finally, neighbouring CLOCK is KIT, which regulates melanocytes73, potentially contributing to the transition from melanic riverine to silvery migratory colouration during smoltification (Fig. 2a). The presence of this assemblage of interrelated genes supports the Omy05 inversion as a supergene with sex-dependent effects on migratory tendency in rainbow trout.
Geographic distribution of the Omy05 rearrangement
The frequency of the rearranged karyotype varied with migratory access in 91 geographically distributed population samples (mean R frequency above versus below barriers, 0.84 versus 0.66, P < 0.001; Fig. 2e, Supplementary Table 4a and Extended Data Fig. 9), as previously observed in the south25, confirming that the link with the migratory trait extends across the species range. However, in the 42 ocean-accessible populations, we observed a strong cline in the R karyotype with both latitude (adjusted R2 = 0.48, P < 0.001; Fig. 2f) and monthly mean ambient temperature (maximum adjusted R2 = 0.61, P < 0.001; Extended Data Fig. 10). These patterns are probably driven by temperature-dependent developmental rates22,74, major quantitative trait loci which overlap the Omy05 rearrangement24,53,74, strongly predict reproductive timing and early male sexual maturation75. Thus, despite the strong influence of the Omy05 double inversion on individual migratory tendency in Big Creek, near the southern extent of the species’ range, this effect is probably reduced where the ancestral, slow-developing karyotype is temperature-limited. In these colder, high-latitude populations, the faster intrinsic developmental rate required to compensate for the effects of decreased temperature may result in positive selection for the rearranged karyotype irrespective of migratory phenotype (Fig. 5). However, anadromy is also rarer at high latitudes, reflecting both the increased food availability in rivers with large salmon runs and increased accumulation of adipose tissue with lower metabolic rates in cooler rivers22,76. These trends reduce the cost of residency for females, suggesting a concomitant decrease in sexual antagonism and that sexual conflict resolution by sex-dependent dominance is dependent on geographical variation in the strength of sex-specific selection1,4,77.
Differences in development, fecundity and relative expression of alternative life-history patterns are influenced by environment (temperature, developmental rate, selection by migration barriers), sex, and Omy05 karyotype, leading to the alternative individual migratory life histories known as rainbow trout and steelhead.
Insights
Inversion supergenes maintained by balancing selection have been associated with the evolution of new dominance patterns, and unlinked modifiers are thought to act epistatically on inversion effects15,44,54. Our results demonstrate sex-dependent dominance reversal of an autosomal inversion contributing to the resolution of sexual conflict over a complex life history trade-off. Sex-dependent dominance of the higher fitness karyotype within each sex provides the conditions for net heterozygote superiority7, allowing independent optimization of the migratory phenotype in both males and females and the maintenance of sexual conflict polymorphisms over a large chromosomal segment. In stark contrast, the Y chromosome lacks signatures typical of sex chromosome evolution, with no evidence for enrichment of male benefit genes, isolating structural rearrangements or an excess of gene loss indicative of degeneracy. These patterns strongly suggest that Y chromosome evolution in rainbow trout is driven primarily by the avoidance of mutation load, with limited capacity to maintain sexual conflict polymorphisms and that the Omy05 rearrangement represents an autosomal alternative to the canonical model of sexual conflict resolution by sex chromosomes.
Linkage is expected to both develop in sexually antagonistic loci and to relax the conditions for maintenance of sexually antagonistic variation because of increased fitness effects78. Such processes can lead to the accumulation of loci with large fitness effects predicted under sexual antagonism79; thus, the sex-dependent dominance reversal of an autosomal supergene we observe in rainbow trout may represent a common mechanism for the maintenance of polygenic sexually antagonistic variation.
In contrast to other inversions underlying alternative reproductive tactics11,12,13,14, lack of homozygous lethality enables the Omy05 rearrangement to purge deleterious mutations from both karyotypes. The inversion complex has been maintained by sexually antagonistic balancing selection for approximately 1.5 million years, and so represents a stable alternative mechanism of sexual conflict resolution that avoids the costs of mutation load accumulation by sex chromosomes. However, this architecture is probably geographically variable, with selection on temperature-dependent developmental rate and the strength of sexual antagonism over alternative migratory tactics changing with latitude. Finally, the maintenance of fitness variation by sexually antagonistic selection has important conservation implications, but geographical variation in the strength of such selection highlights the complexity involved in incorporating adaptive genomic variation into conservation management80,81.
Methods
Genome sequencing and assembly
We sequenced and assembled the rainbow trout genome using DNA from a single homozygous double haploid YY male from the Swanson River (Alaska) clonal line (BioProject no. PRJNA335610) using a complementary combination of inputs, including short-read sequencing technology from Illumina, high-density linkage mapping and the DeNovoMAGIC genome assembly pipeline from NRGene82, as well as long-range data from Dovetail Genomics Chicago library sequencing30 and comparative genomic information from the Atlantic salmon genome27 (Supplementary Information, Section 1.1). To anchor, order and orientate scaffolds into chromosome sequences, we constructed a high-density linkage map using the Lep-MAP software83 (Lep-MAP3 v0.1) from a pedigree of 5,716 fish genotyped with a 57 K SNP array84.
Repeat masked chromosome sequences for rainbow trout were aligned against each other using LASTZ85 (v1.03.73) to identify 98 homeologous blocks originating from the Ss4R (for details, see Supplementary Information, Section 2). Sequence similarity between homeologous sequences was determined in 1-Mb intervals by averaging the local percentage of nucleotide sequence identity using high-scoring segment pairs from LASTZ alignments and presented as a Circos plot86 in Fig. 1.
Whole-genome resequencing
Whole-genome, paired-end sequencing libraries were prepared from genomic DNA extracted from the fin clips of 61 rainbow trout and sequenced using the HiSeq 2000 and 2500 platforms (Illumina), providing an average of 15× and minimum of 8× genome coverage per sample. The sequenced samples included 11 clonal lines from Washington State University, 38 steelhead and resident rainbow trout from wild and hatchery origin populations distributed throughout the native range of the species and 12 fish from the AquaGen rainbow trout aquaculture breeding programme. The origin and geographical location of each sample are given in Supplementary Table 4. The bioinformatics pipeline to map the resequencing data to the reference genome, variant calling and quality filtering of the SNPs has been described previously87. A total of 31,441,105 SNPs were identified genome-wide; the subset of SNPs that mapped to chromosome Omy05 was further used for population genetic and functional analyses as described in the ensuing sections.
Orthologue identification
Orthologues were first identified using OrthoFinder88 (v1.1.9) with protein sequences of two mammalian species (mouse and human) and eight teleost species (zebrafish, medaka, stickleback, northern pike, coho salmon, rainbow trout, European grayling and Atlantic salmon) as the input. Gene annotations for mouse, human, zebrafish, medaka and stickleback were downloaded from Ensembl v86. For northern pike and Atlantic salmon, we used the NCBI RefSeq annotations for assembly versions ASM72191v2 and ICSASG_v2, respectively. For coho salmon, we used TransDecoder v2.0.1 (https://github.com/TransDecoder/TransDecoder/wiki) to predict protein sequences based on a de novo transcriptome assembly. Rainbow trout gene annotations were based on an in-house annotation pipeline described previously and used for Atlantic salmon27.
For each orthogroup, protein sequences were aligned using MAFFT v7.310 and protein trees were estimated using FastTree v2.1.8. Orthogroup protein trees containing duplication nodes ancestral to all teleosts or vertebrates were then further partitioned into smaller clan trees using an in-house R function (available from https://gitlab.com/sandve-lab/salmonid_synteny/blob/master/clanfinder_function.R). New codon alignments for the orthogroups, including those converted to ortho clans with reduced gene tree complexity, were made using Pal2Nal v14.0 to convert protein alignments into CDS alignments. These nucleotide alignments were then used to re-estimate a final set of orthologue gene trees.
Generation of time-calibrated Bayesian Evolutionary Analysis Sampling Trees (BEAST) for genes on Omy05
For each gene on Omy05, we identified the corresponding gene tree and performed a series of filtering steps to retain only orthogroups for which we had high-confidence classification of both orthologues and duplicates originating from the salmonid whole-genome duplication. This was ensured by filtering according to gene tree topologies using the following criteria: (1) salmonid tips had to be monophyletic; (2) gene tree phylogenies had to conform to the species phylogeny for non-salmonid taxa (after rooting in the most distant salmonid outgroup); (3) the gene tree had to have retained all duplicates from the salmonid whole-genome duplication (Ss4R); (4) the topology of Ss4R duplicates had to conform to the salmonid species phylogeny.
After identifying high-confidence gene orthologue groups for rainbow trout genes on Omy05, we made sequence alignments for these orthologue groups using both CDS and whole genes containing introns. Coding regions (CDS and genes) were extracted from the whole-genome sequence VCF file using VCFtools v0.1.14 and then refilled to sequence using the vcf2fasta function in FreeBayes89 (v1.1.0) using gene FASTA files created with SAMtools90 (v1.3). Before alignment, we swapped out the single rainbow trout nucleotide sequence from Omy05 with two sequences corresponding to the two inversion haplotype sequences. Sequences from each orthogroup were then aligned with MAFFT using default parameters; alignments were trimmed using gBlocks91 (v0.91b).
Finally, BEAST92 (v1.7.5) was run on each alignment (CDS and gene) which had >0 base pair differences between the inversion haplotypes using the parameters chainLength = 10000000, storeEvery = 5000, a Yule-model of speciation, the Hasegawa–Kishino–Yano substitution model and a relaxed molecular clock. For priors we assumed: (1) the duplicate salmonid gene lineage including the Omy05 haplotypes to be monophyletic and with a divergence time of 20 Ma (log-normal distribution with an s.d. = 0.1); and (2) northern pike and all salmonids to be monophyletic and diverging 125 Ma (log-normal distribution with an s.d. = 0.1).
Sex chromosome evolution
Between-sex divergence (F ST)
Between-sex Weir and Cockerham’s FST was calculated for each SNP from the whole-genome resequence dataset and also separately for wild individuals, pooled across populations, using VCFtools. The inclusion of YY double-haploid males increased the peak height in the sequence surrounding the sdY location, but this was also the highest between-sex FST peak in wild individuals. Between-sex FST was plotted for Omy29 and compared with the linkage map for this chromosome (see earlier).
Coverage
Genotypic sex was determined by coverage of the sdY-containing scaffold, KJ851798.1:20860–23610, where samples with a coverage of zero were typed as female. Samples were processed using the SpeedSeq pipeline93 (git/0238d5c) and aligned with BWA-MEM94 v.0.7.10-r789. Coverage data were calculated using Mosdepth95 v.0.2.1, both for the whole genome in 500-bp windows and for all exons from the RefSeq100 Omyk_1.0 annotation. For Mosdepth, the mapping quality threshold was set to 10. Log2 coverage was plotted and the significance of the difference in coverage was calculated by a t-test.
Expression (RNA sequencing (RNA-seq))
RNA-seq data from 15 tissues (spleen, kidney, gill, head kidney, skin, intestine, liver, red muscle, white muscle, brain, fat, stomach, pineal gland, oocyte and testis) were downloaded from the Sequence Read Archive (SRA). Quantification of RNA-seq data was performed using Kallisto96 v.0.44.0 with 30 bootstrap samples. We defined testis and oocyte genes as those expressed maximally in the testes and oocytes, respectively, with a TPM > 5. Enrichment of these sex-specific genes on the Y chromosome (Omy29) was tested with Fisher’s exact tests.
Gene gain and loss
The gene content of the orthologous rainbow trout Omy29 and coho Oki29 chromosomes were compared using a combination of gene orthology relationships determined by OrthoFinder (https://doi.org/10.1186/s13059-015-0721-2), the orthologue relationships outlined earlier and protein BLAST. We used BLAST to increase the number of genes with defined orthologue relationships between the two chromosomes. Where the matching gene mapped to an unmapped scaffold and the duplicate mapped to the homeologue of chromosome 29 in the other species, we assumed the unmapped copy belonged on chr29; this means that we may have missed some translocations away from chr29. However, since there were more unmapped chr29 genes in coho than in rainbow trout, we considered it conservative for understanding gene content evolution in rainbow trout.
Genome sequence synteny
Rediploidization of salmonid genomes has progressed through the rearrangement and fission/fusion of large chromosomal blocks with identifiable syntenic relationships of these blocks among species27. Large-scale chromosomal synteny was determined through the comparison of published restriction site-associated DNA sequencing (RAD-seq) linkage maps for coho97, Chinook98, chum99 and sockeye salmon100, and genome sequences available for Arctic char (GCF_002910315.2), coho (GCF_002021735.1) and Atlantic salmon (GCF_000233375.1).
Field sampling and capture–recapture experiment
Fieldwork
The study was conducted in a natural population of O. mykiss in Big Creek, a small (58 km2) coastal watershed along the central California coast in Monterey County, California, USA. Individuals in this population have free access to migrate to the ocean, but many mature residents remain in the creek101. We non-lethally captured, weighed, measured and took caudal fin samples from more than 2,600 individuals in approximately 1,900 m of contiguous stream habitat starting at the Pacific Ocean entry of Big Creek between May 2006 and October 2009. All fish >100 mm fork length were injected with unique passive integrated transponder tags before release at the capture location. Tagged fish were available for detection by a continuously active in-stream fixed antenna approximately 70 m upstream from the ocean that recorded the date and time of migration by detected individuals between May 2006 and October 2012. Due to tag size limitations, only individuals >100 mm fork length received a tag (23-mm half duplex) detectable by the antenna. The length and weight assigned to tagged fish detected migrating from the stream were based on measurements taken at their last physical capture, typically the autumn before migration. Analysis of antenna detections considered the subset of fish (n = 887) that were (1) tagged with passive integrated transponder tags detectable by the antenna, (2) captured ≥100 m upstream from the antenna array near the stream mouth and (3) successfully genotyped for Omy05 and OmyY1 sex identification loci. From this group, ‘migrants’ were defined as individuals detected at least once by the in-stream antenna that (1) had no subsequent capture or detection, (2) were at large <730 d between their last capture and final detection and (3) were detected at the array <15 d in total and over a span of <60 d. These criteria were used to distinguish detections of true migrants (that is, smolts; n = 331) from detections of tags no longer in live fish (for example, following death or predation of a tagged fish; n = 68). The remaining 488 individuals were never detected by the in-stream fixed antenna, so are assumed to be either non-migratory residents or in-stream mortalities.
SNP genotyping
Genomic DNA was extracted from the fin clips of all fish sampled in Big Creek and genotyped at 95 SNPs102 using TaqMan assays (Applied Biosystems) on 96.96 Dynamic SNP Genotyping Arrays with the EP1 Genotyping System (Fluidigm Corporation). An additional TaqMan assay was designed around a Y chromosome-linked gene probe103 and an invariant autosomal control gene and was used to determine genetic sex. Two negative (no DNA) controls were included in each array and genotypes were called using SNP Genotyping Analysis Software v3.1.1 (Fluidigm Corporation). Two loci located within the Omy05 rearrangement, Omy_114448-87 and Omy_121006-131, showed strong linkage disequilibrium with each other and with other loci located within the Omy05 rearrangement25. In Big Creek, these two loci are in near-perfect disequilibrium, with 97.2% identical genotypes. We designated the C and T bases at SNP Omy_114448-87 as A (ancestral/anadromous) and R (rearranged/resident), respectively for the Omy05 rearrangement. The remaining 92 non-Omy05 loci were used for population genetic and kinship analyses.
Capture–recapture modelling
The effects of Omy05 inversion haplotype on migratory behaviour were investigated using a channel-spanning passive integrated transponder tag in-stream antenna approximately 70 m upstream from the ocean. We predicted that the probability of detecting anadromous tagged fish would be highest for fish tagged near the size that migratory steelhead smolts typically emigrate from freshwater104 (approximately 150 mm fork length). Detection probability should be lower for anadromous fish sampled at a smaller size, since many will not survive to emigrate, and also for larger fish, since they are presumably maturing to remain as residents. Fish with genotypes favouring residency may also be observed at the antenna, but we expected this probability to show no peak near 150 mm fork length. To test these hypotheses, we used generalized additive models to estimate the probability of detection at the downstream antenna as a function of length at last capture and release, with Omy05 genotype and sex as categorical covariates, using the mgvc package v.1.8–4 in the statistical programming language R v.3.1.3. Because earlier work showed that the sex ratio of fish smaller than 150 mm was near 50:50 while larger size classes are enriched in males101, we expected that there should be an interaction between sex and genotype. Therefore, we tested seven models capable of accommodating increasing variability. The simplest model assumed that emigration probability is only a smooth function of length at tagging and release. Slightly more complex models included effects of sex and genotype separately (two and three smooth functions, respectively). The final four models treated the sex and genotype interaction differently, allowing for partial dominance (six smooth functions, one for each sex and genotype combination), anadromous or resident dominance or sex-dependent dominance reversal, with anadromy dominant for females and residency dominant for males. The best model according to the AIC included sex-dependent dominance reversal, with an AIC = 4.90 lower than the next highest-ranked model (Supplementary Table 5).
Geographic survey of Omy05 variation
Omy05 SNP populations survey
Tissue samples (approximately 1–3 mm2) or genomic DNA were obtained from 63 natural origin O. mykiss populations (n = 1,592 individuals) ranging from the Kamchatka Peninsula, Russia to southern California, USA. Also, 94 individuals sampled from 4 steelhead hatchery broodstock and 141 individuals sampled from 6 rainbow trout hatchery strains were genotyped (Supplementary Table 4). These samples were all genotyped using a separate panel of 86 SNPs distributed on chromosome Omy05, 55 SNP assays from Pearse et al.25 and 31 developed by Miller et al.74. Eight loci were outside the inversion region and 3 failed to amplify, leaving 75 SNPs retained for analyses. Allele frequencies were assessed using the R package adegenet v.1.3–4 (ref. 105). Linkage disequilibrium was estimated with the allelic correlation coefficient R2 (refs. 106,107) using the R package genetics108. The number of pairwise linkage associations above a critical value of 0.95 were counted for each population. Because pairwise R2 values cannot be estimated in populations where either locus is monomorphic, we divided the number of values above the critical value by the total number of R2 values calculated for each population to obtain a weighted value. We used level plots in R to visualize the allele frequency and linkage disequilibrium data.
Of the 1,592 individuals examined, 286 fish from 33 sampling locations were homozygous AA and 846 fish from 57 sampling locations were homozygous RR. Population level trees were generated with poppr109 v.2.3.0 in R v.3.3.1 with Rstudio v.1.0.136 with the aboot command. Chord distances between populations110 were calculated and a tree constructed with the Neighbor-Joining algorithm111. The confidence of inferred relationships was evaluated with 1,000 bootstrap replicates.
Finally, the SNP population survey dataset was combined with additional data on inversion frequencies for a combined total of 2,249 individuals from 91 populations (Supplementary Table 4.1). For all data sources, inversion frequency was inferred based on common SNPs that perfectly or nearly perfectly identified the inversion haplotype.
Inversion frequency as a function of latitude
A plot of latitude (x axis, WGS84) and inversion frequency (y axis) was generated by filtering inversion frequency data for those samples from North America taken below migration barriers and without known hatchery introgression (Fig. 2 and Supplementary Table 4). Graphics and data analysis occurred in R v.3.3.1 in Rstudio v.1.0.136 with the ggplot2 package v.2.2.1. For each estimate of inversion frequency, the s.e.m. was calculated with P equal to the frequency of the inversion in the sampled population and n equalling the number of individuals sampled from that population. To the plot of latitude and inversion frequency, a weighted least squares regression line was added from a model with inversion frequency dependent on the latitude of the sampled population weighted by the sample size of the population. The resulting model (inversion frequency = 0.04 × (latitude) – 1.21) has an adjusted R2 of 0.51.
Inversion frequency as a function of temperature
Temperature data were obtained for each month of the year from the WorldClim112 dataset v.1.4. The dataset represents current temperatures taken from interpolations of observed data from 1960 to 1990 within 30 arc-second squares. Temperature data were extracted with the raster package v.2.5–8 in R. A bilinear interpolation was implemented to average between the 4 nearest 30 arc-second squares to the collection location. As with the inversion frequency as a function of latitude, a weighted least squares regression weighted by sample size was conducted for each month of the year and the resulting adjusted R2 was retained for each subplot (Extended Data Fig. 10).
Animal use
Double-haploid lines were sampled according to our Standard Operating Procedures: Care and Use of Research Animals, protocol no. 114, US Department of Agriculture (USDA)-Agricultural Research Service (ARS), National Center for Cool and Cold Water Aquaculture. This protocol was reviewed and approved by the institutional animal care and use committee on 2 December 2016. All wild animal samples were collected following National Marine Fisheries Service (NMFS) institutional protocols and approvals, including Endangered Species Act Section 10 permit no. 1044-M4.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The reference genome assembly: GenBank Assembly Accession GCA_002163495.1, RefSeq Assembly Accession GCF_002163495.1. Raw sequence data used for the genome assembly: NCBI SRA Accession SRP086605 (Project ID: Project PRJNA335610). Raw sequence data used for whole-genome resequencing: NCBI SRA Accession SRP107028 (Project ID: PRJNA386519). New RNA-seq data generated for the genome annotation: NCBI SRA Accession SRP102416 (Project ID: PRJNA380337). Additional sequence data used for the NCBI RefSeq annotation are listed and described at https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Oncorhynchus_mykiss/100/. Raw sequence data used for generating RAD SNP markers that were used for anchoring assembly scaffolds and contigs to chromosomes: USDA: NCBI SRA Accession SRP063932 (Project ID: PRJNA295850); UC Davis: NCBI SRA Accession SRP141092 (Project ID: PRJNA450873). NMFS data and analysis can be found at https://github.com/eriqande/Pearse_etal_NEE_NMFS_Data_Analysis.
Change history
09 December 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Connallon, T. The geography of sex-specific selection, local adaptation, and sexual dimorphism. Evolution 69, 2333–2344 (2015).
Lande, R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34, 292–305 (1980).
Mank, J. E. Population genetics of sexual conflict in the genomic era. Nat. Rev. Genet. 18, 721–730 (2017).
Barson, N. J. et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528, 405–408 (2015).
Bachtrog, D. et al. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899 (2014).
Rice, W. R. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41, 911–914 (1987).
Fry, J. D. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution 64, 1510–1516 (2010).
Cavoto, E., Neuenschwander, S., Goudet, J. & Perrin, N. Sex-antagonistic genes, XY recombination and feminized Y chromosomes. J. Evol. Biol. 31, 416–427 (2018).
Spencer, H. G. & Priest, N. K. The evolution of sex-specific dominance in response to sexually antagonistic selection. Am. Nat. 187, 658–666 (2016).
Grieshop, K. & Arnqvist, G. Sex-specific dominance reversal of genetic variation for fitness. PLoS Biol. 16, e2006810 (2018).
Küpper, C. et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83 (2016).
Lamichhaney, S. et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 48, 84–88 (2016).
Tuttle, E. M. et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26, 344–350 (2016).
Wang, J. et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493, 664–668 (2013).
Le Poul, Y. et al. Evolution of dominance mechanisms at a butterfly mimicry supergene. Nat. Commun. 5, 5644 (2014).
Llaurens, V., Whibley, A. & Joron, M. Genetic architecture and balancing selection: the life and death of differentiated variants. Mol. Ecol. 26, 2430–2448 (2017).
Blaser, O., Neuenschwander, S. & Perrin, N. Sex-chromosome turnovers: the hot-potato model. Am. Nat. 183, 140–146 (2013).
Lubieniecki, K. P. et al. Genomic instability of the sex-determining locus in Atlantic Salmon (Salmo salar). G3 (Bethesda) 5, 2513–2522 (2015).
Cavileer, T. D., Hunter, S. S., Olsen, J., Wenburg, J. & Nagler, J. J. A sex-determining gene (sdY) assay shows discordance between phenotypic and genotypic sex in wild populations of Chinook salmon. Trans. Am. Fish. Soc. 144, 423–430 (2015).
Eisbrenner, W. D. et al. Evidence for multiple sex-determining loci in Tasmanian Atlantic salmon (Salmo salar). Heredity (Edinb) 113, 86–92 (2014).
Perrin, N. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63, 3043–3049 (2009).
Kendall, N. W. et al. Anadromy and residency in steelhead and rainbow trout (Oncorhynchus mykiss): a review of the processes and patterns. Can. J. Fish. Aquat. Sci. 72, 319–342 (2015).
Ohms, H. A., Sloat, M. R., Reeves, G. H., Jordan, C. E. & Dunham, J. B. Influence of sex, migration distance, and latitude on life history expression in steelhead and rainbow trout (Oncorhynchus mykiss). Can. J. Fish. Aquat. Sci. 71, 70–80 (2013).
Nichols, K. M., Edo, A. F., Wheeler, P. A. & Thorgaard, G. H. The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genetics 179, 1559–1575 (2008).
Pearse, D. E., Miller, M. R., Abadía-Cardoso, A. & Garza, J. C. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proc. Biol. Sci. 281, 20140012 (2014).
Berthelot, C. et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5, 3657 (2014).
Lien, S. et al. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205 (2016).
Macqueen, D. J. & Johnston, I. A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. Biol. Sci. 281, 20132881 (2014).
Hardie, D. C. & Hebert, P. D. The nucleotypic effects of cellular DNA content in cartilaginous and ray-finned fishes. Genome 46, 683–706 (2003).
Putnam, N. H. et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26, 342–350 (2016).
Phillips, R. B. et al. Assignment of rainbow trout linkage groups to specific chromosomes. Genetics 174, 1661–1670 (2006).
Yano, A. et al. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22, 1423–1428 (2012).
Yano, A. et al. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male‐specific Y‐chromosome sequence in many salmonids. Evol. Appl. 6, 486–496 (2013).
Roberts, R. B., Ser, J. R. & Kocher, T. D. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326, 998–1001 (2009).
van Doorn, G. & Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909–912 (2007).
Vicoso, B., Kaiser, V. B. & Bachtrog, D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl Acad. Sci. USA 110, 6453–6458 (2013).
Kirkpatrick, M. & Guerrero, R. F. Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197, 531–541 (2014).
Phillips, R. B. et al. Characterization of the OmyY1 region on the rainbow trout Y chromosome. Int. J. Genomics 2013, 261730 (2013).
Phillips, R. B. et al. Recombination is suppressed over a large region of the rainbow trout Y chromosome. Anim. Genet. 40, 925–932 (2009).
Paigen, K. & Petkov, P. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11, 221–233 (2010).
Singer, A. et al. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160, 649–657 (2002).
Sutherland, B. J. G. et al. Salmonid chromosome evolution as revealed by a novel method for comparing RADseq linkage maps. Genome Biol. Evol. 8, 3600–3617 (2016).
Blackmon, H. et al. Long-term fragility of Y chromosomes is dominated by short-term resolution of sexual antagonism. Genetics 207, 1621–1629 (2017).
Thompson, M. J. & Jiggins, C. D. Supergenes and their role in evolution. Heredity 113, 1–8 (2014).
Hale, M. C., McKinney, G. J., Thrower, F. P. & Nichols, K. M. RNA-seq reveals differential gene expression in the brains of juvenile resident and migratory smolt rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part D 20, 136–150 (2016).
Nakane, Y. et al. The saccus vasculosus of fish is a sensor of seasonal changes in day length. Nat. Commun. 4, 2108 (2013).
Dickmeis, T. et al. Glucocorticoids play a key role in circadian cell cycle rhythms. PLoS Biol. 5, e78 (2007).
Vatine, G., Vallone, D., Gothilf, Y. & Foulkes, N. It’s time to swim! Zebrafish and the circadian clock. FEBS Lett. 585, 1485–1494 (2011).
Yoshitane, H. et al. JNK regulates the photic response of the mammalian circadian clock. EMBO Rep. 13, 455–461 (2012).
Kolodziejczak, M. et al. Serotonin modulates developmental microglia via 5-HT2B receptors: potential implication during synaptic refinement of retinogeniculate projections. ACS Chem. Neurosci. 6, 1219–1230 (2015).
Ori, M., De-Lucchini, S., Marras, G. & Nardi, I. Unraveling new roles for serotonin receptor 2B in development: key findings from Xenopus. Int. J. Dev. Biol. 57, 707–714 (2013).
Hecht, B. C., Campbell, N. R., Holecek, D. E. & Narum, S. R. Genome-wide association reveals genetic basis for the propensity to migrate in wild populations of rainbow and steelhead trout. Mol. Ecol. 22, 3061–3076 (2013).
Nichols, K. M. et al. Quantitative trait loci × maternal cytoplasmic environment interaction for development rate in Oncorhynchus mykiss. Genetics 175, 335–347 (2007).
Kunte, K. et al. doublesex is a mimicry supergene. Nature 507, 229–232 (2014).
Graf, M., Teo Qi-Wen, E.-R., Sarusie, M. V., Rajaei, F. & Winkler, C. Dmrt5 controls corticotrope and gonadotrope differentiation in the zebrafish pituitary. Mol. Endocrinol. 29, 187–199 (2015).
Guo, Y. et al. Molecular cloning, characterization, and expression in brain and gonad of Dmrt5 of zebrafish. Biochem. Biophys. Res. Commun. 324, 569–575 (2004).
Johnsen, H. & Andersen, Ø. Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. The fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505, 221–232 (2012).
Xu, S., Xia, W., Zohar, Y. & Gui, J.-F. Zebrafish dmrta2 regulates the expression of cdkn2c in spermatogenesis in the adult testis. Biol. Reprod. 88, 1–12 (2013).
Aubin-Horth, N., Landry, C. R., Letcher, B. H. & Hofmann, H. A. Alternative life histories shape brain gene expression profiles in males of the same population. Proc. Biol. Sci. 272, 1655–1662 (2005).
Yin, J. et al. Genes and signaling networks regulated during zebrafish optic vesicle morphogenesis. BMC Genomics 15, 825 (2014).
Cimino, I. et al. Novel role for anti-Müllerian hormone in the regulation of GnRH neuron excitability and hormone secretion. Nat. Commun. 7, 10055 (2016).
Cavileer, T., Hunter, S., Okutsu, T., Yoshizaki, G. & Nagler, J. Identification of novel genes associated with molecular sex differentiation in the embryonic gonads of rainbow trout (Oncorhynchus mykiss). Sex. Dev. 3, 214–224 (2009).
von Hofsten, J. & Olsson, P.-E. Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes. Reprod. Biol. Endocrinol. 3, 63 (2005).
Mrosek, N. et al. Transcriptional regulation of adipocyte formation by the liver receptor homologue 1 (Lrh1)-Small hetero-dimerization partner (Shp) network. Mol. Metab. 2, 314–323 (2013).
Hess, J. E., Zendt, J. S., Matala, A. R. & Narum, S. R. Genetic basis of adult migration timing in anadromous steelhead discovered through multivariate association testing. Proc. Biol. Sci. 283, 20153064 (2016).
Taranger, G. L. et al. Control of puberty in farmed fish. Gen. Comp. Endocrinol. 165, 483–515 (2010).
Kling, P. et al. The role of growth hormone in growth, lipid homeostasis, energy utilization and partitioning in rainbow trout: interactions with leptin, ghrelin and insulin-like growth factor I. Gen. Comp. Endocrinol. 175, 153–162 (2012).
Londraville, R. L., Prokop, J. W., Duff, R. J., Liu, Q. & Tuttle, M. On the molecular evolution of leptin, leptin receptor, and endospanin. Front. Endocrinol. (Lausanne) 8, 58 (2017).
Salmerón, C. et al. Effects of nutritional status on plasma leptin levels and in vitro regulation of adipocyte leptin expression and secretion in rainbow trout. Gen. Comp. Endocrinol. 210, 114–123 (2015).
Day, F. R. et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 49, 834–841 (2017).
Talukder, A. H., Li, D.-Q., Manavathi, B. & Kumar, R. Serine 28 phosphorylation of NRIF3 confers its co-activator function for estrogen receptor-α transactivation. Oncogene 27, 5233–5242 (2008).
Li, D. et al. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol. Cell. Biol. 19, 7191–7202 (1999).
Singh, A. P. & Nüsslein-Volhard, C. Zebrafish stripes as a model for vertebrate colour pattern formation. Curr. Biol. 25, R81–R92 (2015).
Miller, M. R. et al. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol. Ecol. 21, 237–249 (2012).
Thrower, F. P., Hard, J. J. & Joyce, J. E. Genetic architecture of growth and early life-history transitions in anadromous and derived freshwater populations of steelhead. J. Fish Biol. 65, 286–307 (2004).
Quinn, T. P. & Myers, K. W. Anadromy and the marine migrations of Pacific salmon and trout: Rounsefell revisited. Rev. Fish Biol. Fish. 14, 421–442 (2004).
Czorlich, Y., Aykanat, T., Erkinaro, J., Orell, P. & Primmer, C. R. Rapid sex-specific evolution of age at maturity is shaped byo genetic architecture in Atlantic salmon. Nat. Ecol. Evol. 2, 1800–1807 (2018).
Patten, M. M., Haig, D. & Ubeda, F. Fitness variation due to sexual antagonism and linkage disequilibrium. Evolution 64, 3638–3642 (2010).
Connallon, T. & Clark, A. G. Balancing selection in species with separate sexes: insights from Fisher’s geometric model. Genetics 197, 991–1006 (2014).
Kardos, M. & Shafer, A. B. A. The peril of gene-targeted conservation. Trends Ecol. Evol. 33, 827–839 (2018).
Pearse, D. E. Saving the spandrels? Adaptive genomic variation in conservation and fisheries management. J. Fish Biol. 89, 2697–2716 (2016).
Hirsch, C. N. et al. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28, 2700–2714 (2016).
Rastas, P., Paulin, L., Hanski, I., Lehtonen, R. & Auvinen, P. Lep-MAP: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29, 3128–3134 (2013).
Palti, Y. et al. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 15, 662–672 (2015).
Harris, R. S. Improved Pairwise Alignment of Genomic DNA. PhD thesis, Pennsylvania State Univ. (2007).
Krzywinski, M. I. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Gao, G. et al. A new single nucleotide polymorphism database for rainbow trout generated through whole genome resequencing. Front. Genet. 9, 147 (2018).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/pdf/1207.3907.pdf (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Chiang, C. et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods 12, 966–968 (2015).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/pdf/1303.3997.pdf (2013).
Pedersen, B. S. & Quinlan, A. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Kodama, M., Brieuc, M. S., Devlin, R. H., Hard, J. J. & Naish, K. A. Comparative mapping between Coho Salmon (Oncorhynchus kisutch) and three other salmonids suggests a role for chromosomal rearrangements in the retention of duplicated regions following a whole genome duplication event. G3 (Bethesda) 4, 1717–1730 (2014).
Brieuc, M. S., Waters, C. D., Seeb, J. E. & Naish, K. A. A dense linkage map for Chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 (Bethesda) 4, 447–460 (2014).
Waples, R. K., Seeb, L. W. & Seeb, J. E. Linkage mapping with paralogs exposes regions of residual tetrasomic inheritance in chum salmon (Oncorhynchus keta). Mol. Ecol. Resour. 16, 17–28 (2016).
Larson, W. A. et al. Identification of multiple QTL hotspots in sockeye salmon (Oncorhynchus nerka) using genotyping-by-sequencing and a dense linkage map. J. Hered. 107, 122–133 (2016).
Rundio, D. E., Williams, T. H., Pearse, D. E. & Lindley, S. T. Male-biased sex ratio of nonoanadromous Oncorhynchus mykiss in a partially migratory population in California. Ecol. Freshw. Fish 21, 293–299 (2012).
Abadía-Cardoso, A., Anderson, E. C., Pearse, D. E. & Garza, J. C. Large-scale parentage analysis reveals reproductive patterns and heritability of spawn timing in a hatchery population of steelhead (Oncorhynchus mykiss). Mol. Ecol. 22, 4733–4746 (2013).
Brunelli, J. P., Wertzler, K. J., Sundin, K. & Thorgaard, G. H. Y-specific sequences and polymorphisms in rainbow trout and Chinook salmon. Genome 51, 739–748 (2008).
Bond, M. H., Hayes, S. A., Hanson, C. V. & MacFarlane, R. B. Marine survival of steelhead (Oncorhynchus mykiss) enhanced by a seasonally closed estuary. Can. J. Fish. Aquat. Sci. 65, 2242–2252 (2008).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Hill, W. G. & Robertson, A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231 (1968).
Pritchard, J. K. & Przeworski, M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69, 1–14 (2001).
Warnes, G. & Leisch, F. The genetics Package: Population Genetics. R package version 1.2.0 (2005).
Kamvar, Z. N., Tabima, J. F. & Grünwald, N. J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014).
Cavalli-Sforza, L. L. & Edwards, A. W. F. Phylogenetic analysis: models and estimation procedures. Evolution 21, 550–570 (1967).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Acknowledgements
We thank H. Fish, K. Pipal and many others for help with fieldwork; V. Apkenas, A. Carlo and E. Campbell for assistance with data collection and analysis; and M. Readdie and F. Aryas for support at the University of California Landels-Hill Big Creek Reserve. Samples and data for the geographic survey were provided by M. Ackerman, S. Lewis, S. Narum, K. Nichols, S. Northrup (Freshwater Fisheries Society of British Columbia), E. Taylor (University of British Columbia), D. Teel and K. Warheit. Compute Canada provided the computing resources used in repeat annotation and analysis. We thank R. Long and K. Shewbridge for their help in DNA sample preparation for sequencing and genotyping and in the preparation of RAD-seq libraries, and K. Martin and Troutlodge for the permission to use samples from their germplasm for genotyping. We also thank the Genomics Core at Washington State University, Spokane, the University of Idaho Genomics Core and the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley for performing DNA library preparation and clonal lines’ resequencing. The genome resequencing of the Whale Rock female clonal line was conducted in collaboration with M. Garvin, Oregon State University. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer. This project was supported by funds from the USDA-ARS (in-house project nos. 1930-31000-009 and 8082-31000-012). DH clonal line resequencing was supported by an Agriculture and Food Research Initiative Competitive Grant (no. 2015-07185) from the USDA National Institute of Food and Agriculture and by an NRSP8 Aquaculture Genome funding seed grant to M.G. and G.T. The whole-genome resequencing data provided by K. Naish was obtained from a project supported by an Agriculture and Food Research Initiative Competitive Grant (no. 2012-67015-19960) from the USDA National Institute of Food and Agriculture. Funding for bioinformatics and statistical support at CIGENE (Norwegian University of Life Sciences) was provided by NFR grants (nos. 208481, 226266 and 275310). Bioinformatics analyses were performed using resources at the Orion Computing Cluster at CIGENE, with storage resources provided by the Norwegian National Infrastructure for Research Data (project no. NS9055K). We acknowledge the help of S. Karoliussen and M. Arnyasi at CIGENE for generating rainbow trout genotypes and M. Baranski for work on the genetic linkage maps. C. R. Primmer and K. Nichols provided valuable comments on the draft manuscript.
Author information
Authors and Affiliations
Contributions
S.Lien, Y.P. and A.G.H. co-conceived the genome assembly project. G.H.T. provided the Swanson clonal line for the reference genome. Y.P. and T.M. contributed SNP chip and RAD SNP genotype data for the linkage analysis. S.Lien and T.M. performed the linkage analyses. T.N. and S.Lien refined the assembly and built the chromosome sequences. K.B., G.B.-Z., D.S.-T. and O.B. designed and conducted the DeNovo MAGIC genome assembly of the Swanson clonal line Illumina sequence data. M.R.M. and L.C. provided RAD SNP data and linkage information for chromosome anchoring of the assembly scaffolds and contigs. D.E.P. and J.C.G. contributed the Dovetail sequence data for the genome assembly. G.G. incorporated the Dovetail sequence data for bridging and combing genome assembly scaffolds. S.Lien, Y.P., G.H.T., B.F.K., N.J.B. and D.E.P. designed the whole-genome resequencing study. G.H.T., B.F.K., S.Liu, K.K., K.A.N., M.S.O.B. and T.M. contributed samples, data and/or analysis to the resequencing study. D.R.M. and B.F.K. created and annotated the repeat library and performed the Tc1-mariner analysis. G.G. performed the bioinformatics analyses on the SNP chip genotype and RAD sequence data. N.J.B., G.G. and M.A.C. analysed the whole-genome resequencing data. S.Lien produced data and completed the comparative genomic analyses. N.J.B., M.K., T.N. and S.Lien produced and analysed the genotype data. M.M., M.K., T.N. and S.Lien made the draft nanopore genome assembly. N.J.B. and M.A.C. performed the population genomic analysis of the inversions using resequence data and analysed the gene content of the inversions. N.J.B. and T.N. performed the analysis of sex chromosome evolution. S.R.S., M.K. and T.N. generated RNA data. S.R.S. generated the orthogroup gene trees. N.J.B. and S.R.S. dated the inversions on Omy05. D.E.R., T.H.W., D.E.P., E.C.A., J.C.G. and S.T.L. conceived, designed and conducted the Omy05 capture–recapture field experiment, and D.E.R., E.C.A., D.E.P. and S.T.L. analysed the data. A.A.-C., J.C.G. and D.E.P. conceived, designed and conducted the SNP populations survey. E.B.R. and B.F.K. contributed additional data, and A.A.-C., E.C.A. and M.A.C. analysed the data. N.J.B., S.Lien, E.C.A., M.A.C., S.T.L., D.E.P. and B.F.K. created the figures. D.E.P., N.J.B., Y.P. and S.Lien wrote the paper with input from all authors. All authors read, commented on and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Animal use All animal handling was conducted in accordance with approved institutional guidelines.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Activity periods, abundance, and historical proliferation of Tc1-Mariner families in Atlantic salmon, rainbow trout, and Chinook salmon.
a. Lower sequence similarity between family members indicates a more ancient family. Activity and abundance are generally consistent among the three species until the time corresponding to ~ 93% sequence similarity, after which substantial differences in activity have occurred in concert with salmonid lineage divergence. Tc1-Mariner families displayed were identified in Atlantic salmon and rainbow trout and occupied at least 0.1% of the genome in one of the three species. b. Stacked density plot of pairwise similarity between Tc1-Mariner family members. The large initial peak with a maxima at ~86% corresponds roughly to the same time that the salmonid-specific whole-genome duplication took place. In the time corresponding to more than 93% similarity, differences in activity begin to appear between Atlantic salmon and rainbow trout in accordance with their ancestral divergence (compare to Fig. 3a in Lien et al, 2016).
Extended Data Fig. 2 High-density linkage maps describing characteristic sex-specific recombination patterns and resolving variable chromosome numbers associated with centric fusions or fissions in rainbow trout.
a. Linkage map for the metacentric rainbow trout chromosome 8 (Omy08) demonstrating male recombination strongly localized towards both telomeres and female recombination repressed at the centromere. b. Linkage map for the metacentric rainbow trout chromosome 2 (Omy02) demonstrating elevated male recombination towards the telomeric region at the q-arm but repressed recombination at the p-arm typified by showing high sequence similarity to Omy03p. c. Linkage map for the acrocentric rainbow trout chromosome 29 (Omy29) with the sex-determining gene sdY located around 5 Mb demonstrating repressed male recombination for most of the chromosome except the telomeric region. d.-f. Rainbow trout chromosomes with variable chromosome numbers associated with centric fusions or fissions. Gaps in the linkage map at the centromere of Omy04, Omy14 and Omy25 are caused by fissions splitting metacentric chromosomes into two acrocentric chromosomes in some families.
Extended Data Fig. 3 Large polymorphic chromosomal rearrangements in the rainbow trout genome.
a. Ancestral (A) versus rearranged (R) chromosomal positions showing structure of inversion complexes on Omy05 and Omy20. b. Genetic linkage maps constructed for parents with alternate Omy05 and Omy20 haplotypes. Red line; female map for homozygous ancestral (AA) parents. Blue line; male map for homozygous ancestral (AA) parents. Orange line; female map for homozygous rearranged (RR) parents with marker order as in ancestral rearrangement. Green line; female map in heterozygous parents (AR).
Extended Data Fig. 4 Chromosomal rearrangements on rainbow trout chromosome 5 (Omy05) and conserved synteny with other salmonid species.
a. Ancestral (A) or rearranged (R) haplotypes on Omy05 characterized by two adjacent inversions of 22.83 and 32.94 Mb. B. Linkage map of recombination on chromosome Omy05 in males (green) and females (red). c.The alignment of Omy05 with the rest of the rainbow trout genome assembly show conserved collinear blocks of homeology with Omy12p, Omy29, Omy01p and Omy04p. d. The alignment of Omy05 with Atlantic salmon genome assembly (GCF_000233375.1) identifies highly conserved synteny with salmon chromosomes 1 and 10 (Ssa01qb and Ssa10qa). e. Alignment with the Arctic char genome (GCF_002910315.2) detects highly conserved synteny with char chromosomes 4 and 16 (Sal04 and Sal16). f. Alignment of Omy05 with coho salmon chromosome sequences (GCF_002021735.1) reveals conserved synteny with chromosomes 23 and 13 (Oki23 and Oki13). g. Comparison of Omy05 with RAD-based linkage maps for other Pacific salmon reveal a smaller fragment at centromere of Omy05 which is rearranged in coho, Chinook, chum and sockeye compared to rainbow trout, Arctic char and Atlantic salmon, and a larger rearrangement that differentiate coho and chinook from the other salmonid species.
Extended Data Fig. 5 Characterization of Omy05 double-inversion break points.
a. Contigs of the ancestral type spanning the three inversion break points on Omy05. The contigs were generated by long-read nanopore sequencing of a fish known to be homozygous for the ancestral configuration of the double inversion. b. Structure of the two variants of the double-inversion on Omy05 categorized as ancestral or rearranged. c. Scaffolds of the rearranged type spanning the three inversion break points on Omy05. The scaffolds were generated from the Swanson doubled haploid line known to be homozygous for the rearranged type of the double-inversion. Tables in the lower part of the figure list SNPs flanking the inversion break points, with positions in ancestral contigs or rearranged scaffolds, respectively, as well as boarders for the three break points in the ancestral type of the double-inversion.
Extended Data Fig. 6 Age estimates of the Omy05 inversion complex.
a. Age estimates from individual gene and CDS across the inversion showing lack of a strong pattern associated with inversion break points. b. Boxplot of age estimates for all CDS and genes that passed filtering, and the difference between the two estimates. Older estimates and wider confidence intervals are obtained from CDS than those based on gene sequences. c. Plot of the number of base differences between haplotypes for gene and CDS alignments and its effect on age estimation. Despite potentially having more variants because of the inclusion of introns, the gene alignments actually have fewer base differences per haplotype on average owing to the removal of poor quality intronic alignments by Gblocks. As a result, the CDS based alignments provide more informative alignments for dating the inversion complex. d. Estimate of inversion age in 10 Mb windows across the inversion complex for CDS estimates.
Extended Data Fig. 7 Full model results from antennae detection emigration model.
Full model results from antennae detection emigration model in Big Creek, showing mean (solid lines) and SD (dotted lines) of differential size-dependent migration of males and females with AR and AA, and RR Omy05 genotypes, with smoltification peaking at ~150 mm.
Extended Data Fig. 8 Linkage maps and conserved synteny with other salmonids for rainbow trout chromosome 29 (Omy29).
a. Sex-specific linkage maps for Omy29 (red dots; female, blue dots; male) show repressed recombination in males for the majority of Omy29. Genetic differentiation between males and females (Fst) is peaking at the sdY locus located at 5 Mb (yellow triangle) but is also elevated in the region between sdY and the centromere (Black Dot at 0 Mb). b. Regions of the rainbow trout genome homeologous with Omy29. Comparative genome sequence maps show highly conserved synteny with c. Atlantic salmon chromosome 11qb (ssa11qb) and d. Arctic char chromosome Sal05. e. Comparative mapping with coho salmon genome sequence reveals two larger rearrangements, one at 10 Mb and one at 30 Mb. f. Comparison of Omy29 with RAD-based linkage maps for other Pacific salmon show that the rearrangement at 10 Mb is conserved among coho, chinook, chum and sockeye and differentiated from rainbow trout and Atlantic salmon.
Extended Data Fig. 9 Genetic Diversity and Interrelationships of AA and RR Omy05 haplotypes.
Principal component (PC) analysis of genetic diversity estimated from (a) whole-genome sequence data genome-wide and, (b) within the chromosome Omy5 rearrangement. For both A and B, a random subset of 40,000 SNPs were examined from homozygous AA and RR individuals of known geographic origin and plotted with the principal components containing the most variance. Homozygous AA individuals are plotted as blue circles with homozygous RR individuals represented by red triangles, with numbers indicating geographic locations corresponding to Fig. 2d and Supplementary Information Table 4. While geographic structuring is apparent in the genome-wide SNP dataset, the inversion region separates clearly between RR and AA types with PC1 (28.76% of variation), and the subsequent second PC (7.49% of variation) corresponding to diversity within AA types. Similarly, population-level Neighbor-Joining trees of AA individuals and RR individuals from SNP survey data of homozygous AA individuals (c) and homozygous RR individuals (d) from sampled populations are depicted in a population level tree generated through chord distances. Support values for nodes with > 50% bootstrap support generated from 1,000 bootstrap replicates are indicated. Population numbers correspond to Fig. 2d and Supplementary Information Table 4.
Extended Data Fig. 10 Inversion Frequency as a Function of Mean Monthly Temperature.
For each month of the year below barrier North American populations of rainbow trout are plotted as mean monthly temperature (x – axis) and inversion frequency (y – axis). Points are sized proportionally to sample size (N). A weighted least squares regression is depicted with the adjusted R2 value for each month of the year.
Supplementary information
Supplementary Information
Supplementary Information and Tables 1–3, 5 and 8.
Supplementary Table 4
Details of all samples used.
Supplementary Table 6
Genes in the Omy05 inversion.
Supplementary Table 7
Homeologous blocks in the O. mykiss genome.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pearse, D.E., Barson, N.J., Nome, T. et al. Sex-dependent dominance maintains migration supergene in rainbow trout. Nat Ecol Evol 3, 1731–1742 (2019). https://doi.org/10.1038/s41559-019-1044-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-1044-6
This article is cited by
-
Strengths and limitations of reduced representation bisulfite sequencing (RRBS) in the perspective of DNA methylation analysis in fish: a case-study on rainbow trout spermatozoa
Fish Physiology and Biochemistry (2024)
-
Genomic selection models substantially improve the accuracy of genetic merit predictions for fillet yield and body weight in rainbow trout using a multi-trait model and multi-generation progeny testing
Genetics Selection Evolution (2023)
-
Genetic architecture of acute hyperthermia resistance in juvenile rainbow trout (Oncorhynchus mykiss) and genetic correlations with production traits
Genetics Selection Evolution (2023)
-
Inversions maintain differences between migratory phenotypes of a songbird
Nature Communications (2023)
-
The roles of sexual selection and sexual conflict in shaping patterns of genome and transcriptome variation
Nature Ecology & Evolution (2023)