Abstract
Biodiversity has always been predominantly microbial, and the scarcity of fossils from bacteria, archaea and microbial eukaryotes has prevented a comprehensive dating of the tree of life. Here, we show that patterns of lateral gene transfer deduced from an analysis of modern genomes encode a novel and abundant source of information about the temporal coexistence of lineages throughout the history of life. We use state-of-the-art species tree-aware phylogenetic methods to reconstruct the history of thousands of gene families and demonstrate that dates implied by gene transfers are consistent with estimates from relaxed molecular clocks in Bacteria, Archaea and Eukarya. We present the order of speciations according to lateral gene transfer data calibrated to geological time for three datasets comprising 40 genomes for Cyanobacteria, 60 genomes for Archaea and 60 genomes for Fungi. An inspection of discrepancies between transfers and clocks and a comparison with mammalian fossils show that gene transfer in microbes is potentially as informative for dating the tree of life as the geological record in macroorganisms.
Similar content being viewed by others
Main
Until Zuckerkandl and Pauling put forth the ‘molecular clock’1 hypothesis, the geological record alone provided the timescale for evolutionary history. Their demonstration that distances between amino acid sequences correlate with divergence times estimated from fossils showed that information in DNA can be used to date the tree of life. Since then, the theory and methodology of the molecular clock have been developed extensively, and inferences from clock analyses (such as the diversification of placentals before the demise of dinosaurs2,3) are hotly debated. Despite these controversies, combining information from rocks and clocks is now widely accepted to be indispensable3,4,5, whereby state-of-the-art estimates of divergence times rely on sequence-based relaxed molecular clocks anchored by multiple fossil calibrations. This approach provides information on both the absolute timescale and the relative variation of the evolutionary rates across the phylogeny (Fig. 1a). Yet, because most life is microbial, and most microbes do not leave discernable fossils, major uncertainties remain about the ages of microbial groups and the timing of some of the earliest and most important events in the evolutionary history of life6,7.
a, The geological record provides the only source of information concerning absolute time. That is, the age of the oldest fossil representative of a clade provides direct evidence of its minimum age (for example, the broken line for the blue clade), but inferring maximum age constraints (for example, the broken line for the red clade), and by extension the relative age of speciation nodes, must rely on indirect evidence of the absence of fossils in the geological record5,31,42,43. b, Gene transfers, in contrast, do not carry information on absolute time, but they do define relative node age constraints by providing direct evidence of the relative age of speciation events. For example, the gene transfer depicted by the black arrow implies that the diversification of the blue donor clade predates the diversification of the red clade (that is, node D is necessarily older than node R). Note, however, that the depicted transfer is not informative about the relative age of nodes D′ and R. c, Sequence divergence (here measured in units of expected number of nucleotide substitutions along a strict molecular clock time tree, see Supplementary Information) for 36 mammals2 is correlated (Pearson’s R2 = 0.664, P < 0.003) with age estimates based on the fossil record (ages corresponding to the time of divergence in million years (Myr)). d, A similar relationship can be seen for gene transfer-based relative ages by plotting the sequence divergence (measured similar to c) against the relative age of ancestral nodes for 40 cyanobacterial genomes (Spearman’s rank correlation ρ = 0.741, P < 10−6) inferred by the MaxTiC algorithm25.
In addition to leaving only a faint trail in the geological record, the evolution of microbial life has left a tangled phylogenetic signal due to extensive lateral gene transfer (LGT). LGT, the acquisition of genetic material potentially from distant relatives, has long been considered an obstacle for reconstructing the history of life8 because different genetic markers can yield conflicting estimates of the phylogeny of a species. However, it has been previously shown that transfers identified using appropriate phylogenetic methods carry information that can be harnessed to reconstruct a species history9,10,11,12,13,14. This reconstruction is possible because different hypotheses of species relationships yield different LGT scenarios and can therefore be evaluated using phylogenetic models of genome evolution15–19. But, in addition to carrying information about the relationships among species, transfers can carry a record of the timing of species diversification because they have occurred between species that existed at the same time10,20,21. As a consequence, a transfer event can be used to establish a relative age constraint between nodes in a phylogeny independently of any molecular clock hypothesis. That is, the ancestor node of the donor lineage must predate the descendant node of the receiving lineage (Fig. 1b, Supplementary Fig. 8). Below, we show that the dating information carried by transfers is consistent with molecular clock-based estimates of relative divergence times in representative groups from the three domains of life.
Results
We examined genome-scale datasets consisting of homologous gene families from complete genomes in Cyanobacteria (40 genomes22), Archaea (60 genomes11) and Fungi (60 genomes23). For each gene family, we used the species tree-aware probabilistic gene tree inference method called ‘amalgamated likelihood estimation (ALE) undated’22,24 to sample evolutionary scenarios involving events of duplication, transfer and loss of genes conditional on a rooted, but undated species phylogeny and multiple sequence alignment of the family. We recorded the donor and recipient for each transfer, using the frequency with which that transfer was observed in the entire sample to score support. We then used a newly developed optimization method called ‘maximum time consistency’ (MaxTiC)25 (see Methods and Supplementary Information) to extract a maximal subset of consistent transfers that specifies a time order of speciation events in the species tree. We found that the maximal subset of transfers implies a time order of speciations that correlates with the distance between amino acid sequences of extant organisms (Spearman’s ρ = 0.741, P < 10−6; Fig. 1d, Supplementary Fig. 9). A similar correlation (Fig. 1c) can be observed if, following Zuckerkandl and Pauling1, we compare fossil dates and sequence divergence in mammals2 (10 time points, Pearson’s R2 = 0.664, P < 0.003 and Spearman’s ρ = 0.83, P = 0.0056).
We observed a strong correlation between time estimates from MaxTiC and molecular clocks in all our datasets (P < 10−3; Supplementary Figs. 14–16). This result suggests that LGTs indeed carry information on the relative age of nodes in all three domains of life. However, this result is not conclusive because part of the correlation trivially arises from the fact that parent nodes are necessarily both older and more distant to extant sequences than their direct descendants26. To control for this effect, we compared the relative time orders of speciation events inferred from transfers to dates obtained using molecular clocks in the absence of calibrations. As a control for the shape of the tree, we measured the random expectation by sampling chronograms from the prior on divergence times but keeping the species phylogeny fixed (without any sequence information). To compare the dating information from transfers to the information conveyed by fossils, we used the same uncalibrated approach on the same mammalian dataset as above2,27 and derived relative node age constraints from fossil calibrations (see Supplementary Information). For the Bacteria, Archaea and Fungi datasets, we derived relative node age constraints from the maximal consistent subsets of transfers obtained using MaxTiC25. For both fossil-based and transfer-based constraints, we then measured the fraction of constraints that are in agreement with each chronogram. As shown in Fig. 2, both fossil-based and transfer-based constraints agree with uncalibrated molecular clocks significantly more than expected by chance. The observed agreement is robust against the choice of different clock models (Fig. 2), priors on divergence time and models of protein evolution (Supplementary Figs. 17–19). This result demonstrates the presence of a genuine and substantial dating signal in gene transfers.
a, Relative ages derived from 12 fossil calibrations from a phylogeny of 36 extant mammals were compared with node ages sampled from four different relaxed molecular clock models implemented in Phylobayes and with node ages derived from random chronograms, keeping the species phylogeny fixed. b–d, Relative ages derived from gene transfers in Cyanobacteria (b), Archaea (c) and Fungi (d) using the MaxTiC algorithm were compared with estimates from the same five models as in a. For each model and each sampled chronogram, we calculated the fraction of relative age constraints that are satisfied. Each violin plot shows the distribution of the fraction of relative age constraints satisfied by 5,000 sampled chronograms. Inside the violins, boxes correspond to the first and third quartiles of the distribution, while a thick horizontal line corresponds to the median, and the whiskers extend to extrema no farther than 1.5 times the interquartile range. The blue distribution corresponds to random chronograms drawn from the prior with the 95% confidence interval denoted by broken lines. The orange distribution corresponds to the strict molecular clock model, purple to the autocorrelated lognormal model, green to the uncorrelated gamma model and grey to the white-noise model.
Interestingly, the molecular clock models show differences in their agreement with relative time constraints. As expected, the strict molecular clock model generally explores a narrow range of dated trees compared with relaxed clocks. However, on average, chronograms based on the strict molecular clock agree less with relative time constraints than those based on relaxed clock models. This effect is particularly clear in mammals, for which the median fraction of satisfied constraints falls within the 95% confidence interval of the random control (Fig. 2a). This result is caused, in large part, by the accelerated evolutionary rate in rodents being interpreted (in the absence of fossil calibrations) as evidence for an age older than that implied by fossils (Supplementary Fig. 4). The lognormal model is best suited to recover such autocorrelated (for example, clade-specific) rate variations along the tree, and indeed exhibits a median of 100% agreement with fossil-based relative age constraints. The uncorrelated gamma model performs second best, perhaps because it is, in fact, autocorrelated along each branch27. Consistent with this idea, the completely uncorrelated white-noise model fares the worst (Fig. 2a–d). This result is in agreement with previous model comparisons in eukaryotes, vertebrates and mammals27. A similar pattern is apparent when considering LGT-derived relative age constraints in Cyanobacteria, Archaea and Fungi, suggesting strong autocorrelated variation of evolutionary rates in these groups that are best recovered using the lognormal model (Fig. 2b–d).
The motivating principle of the MaxTiC algorithm is that transfers from the maximum consistent set carry a robust and genuine dating signal, while conflicting transfers are likely artefactual. Two lines of evidence suggest that this is indeed the case. First, the agreement of relative time constraints derived from transfers excluded by MaxTiC with the node ranking inferred by uncalibrated molecular clocks tends to be lower than random (Supplementary Fig. 12). Second, while the average sequence divergences for donor clades tend to be higher than for corresponding recipient clades in the set of self-consistent transfers (P < 10−8, one sided t-test for difference greater than zero; Fig. 3), they are lower for those discarded by MaxTiC (P < 10−8, one sided t-test for difference lower than zero; Fig. 3).
For genuine LGTs, the donor lineage must be at least as old as the recipient. As one proxy to investigate whether this is the case for transfers retained by our MaxTiC algorithm, we calculated clade-to-tip distances (see Supplementary Information for details) for the inferred donor and recipient clades for LGTs that were retained and discarded by MaxTiC. In all three datasets, transfers retained by MaxTiC (in red) have the property that donor clades are farther from the tips of the tree than recipient clades, but the opposite pattern is observed for conflicting transfers rejected by MaxTiC (green), consistent with the idea that MaxTiC identifies genuine LGTs.
One obvious difference between the fossil-based and transfer-based relative ages presented in Fig. 2 is that the level of agreement is patently lower for transfer-based relative ages. While in mammals approximately half of the chronograms proposed by the lognormal model agree with 100% of the relative constraints, for other datasets no model reaches 80% agreement. This result indicates that some relative constraints derived from LGT consistently disagree with uncalibrated molecular clock estimates. These disagreements are difficult to interpret because both molecular clocks and our transfer-based inferences may be subject to error; simulations suggest that spurious gene transfer inferences do occur with ALE, albeit at a low rate25 (Supplementary Fig. 23). Nonetheless, the low error rate obtained from simulations suggests that at least some transfers contradicting the molecular clocks are genuine. This result yields the exciting idea of a new source of dating information, independent of and complementary to the molecular clock.
To gain further insight into the robustness of these transfer-based estimates, we evaluated their statistical support from the data. Since MaxTiC yields a fully ordered species tree, the relative age constraints derived from its output are potentially overspecified and include constraints with relatively low statistical support. To ascertain the extent of overspecification, we evaluated the statistical support of relative constraints by taking random samples of 50% of gene families and reconstructing the corresponding MaxTiC 1,000 times (Supplementary Figs. 20–22). We then counted the number of times a constraint was observed. In all datasets, a large majority of constraints were highly supported (found in at least 95% of the replicates), and among these, a significant number (between 20% and 32%) consistently disagreed with molecular clock estimates (see Supplementary Table 2). The strongly supported transfer-based constraints that disagree with the clocks could result from the inability of uncalibrated molecular clock estimates to recover the correct timing of speciations in groups with large variations in the substitution rate over time.
Specifically, LGTs provide strong support for the relatively recent emergence of the Prochlorococcus–Synechococcus clade in Cyanobacteria (blue clade in Fig. 4a, estimated age 0.86 billion years ago (Ga)), irrespective of the uncertainty in the root of Cyanobacteria (see Supplementary Information). Although the Prochlorococcus–Synechococcus clade is inferred to be ancient by three of the four uncalibrated molecular clock models in our study, previous analyses using relaxed molecular clock methods with more extensive species sampling and several fossil calibrations, including fossils dating akinete-forming Cyanobacteria at up to 2.1 Ga28 (green in Fig. 4a, estimated age 1.95 Ga) have consistently dated this clade as younger than most of the rest of cyanobacterial diversity29,30. Prochlorococcus have a known history of genome reduction and evolutionary rate acceleration31, which may lead to artefactually ancient age inferences under uncalibrated molecular clock models, as for rodents (discussed above). This result demonstrates that relative time orders implied by LGT can, like fossils, provide a consistent dating signal that is independent of the rate of sequence evolution.
Five thousand chronograms with a speciation time order compatible with LGT-based constraints were sampled per dataset and calibrated to geological time for Cyanobacteria (a), Archaea (b) and Fungi (c) (for details see Methods). The countinous black line corresponds to the consensus chronogram. Red shading represents the spread of node orders within the sample: nodes are in bright red if there is little or no uncertainty on their order according to LGT, in a light red smear if there is high uncertainty on their order. Dates in units of millions of years ago are provided for clades discussed in the text, which are labelled and shaded. Confidence intervals indicate 95% highest probability density (HPD) of the time calibrated time orders with the exception of nodes, indicated with an asterisk, that had unambiguous calibrated time orders for which the 95% HPD of the corresponding node from Supplementary Figs. 25–27 is given. Supplementary Figs. 1–3 provide the same consensus chronograms with species names at the tips.
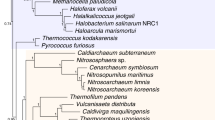
In Archaea, patterns of LGT suggest that several nodes within the Euryarchaeota, including cluster 1 and 2 methanogens (blue and purple clades in Fig. 4 with estimated ages of 3.0 Ga and 2.8 Ga, respectively) are older than both the TACK + Lokiarchaeum clade and DPANN Archaea. The TACK + Lokiarchaeum clade (green clade in Fig. 4) unite Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota with Lokiarchaeum, and have an estimated age of 2.3 Ga. DPANN Archaea (grey in Fig. 4) consist of a genomically diverse group with small cells and genomes, with reduced metabolism suggestive of symbiont or parasite lifestyles, and have an estimated age of 1.8 Ga. The relative antiquity of methanogens is consistent with evidence of biogenic methane at a very early stage of the geological record (~3.5 Ga32), and with another recent analysis that used a single LGT to place the origin of methanogens before the radiation of Cyanobacteria14. These relationships are not recovered by any of the molecular clock models, and suggest that LGT-derived constraints may be highly informative for future dating studies.
The relative order of appearance of archaeal energy metabolisms corresponds to increasing energy yield, with methanogenesis evolving before sulfate reduction, and the oxidative metabolisms of Thaumarchaeota and Haloarchaea evolving most recently. In addition, we find that Ignicoccus hospitalis branches before its obligate parasite Nanoarchaeum (see Supplementary Fig. 2), despite the early divergence of the DPANN clade from other Archaea.
In Fungi, we recover LGTs that provide information on the order of some of the deepest splits. In particular, among crown groups, LGTs indicate that Zoopagomycota33 (blue in Fig. 4, estimated age of 0.71 Ga) diverged earlier than Mucoromycotina, Basidiomycota and Ascomycota (purple, grey and green in Fig. 4, estimated ages of 0.24 Ga, 0.64 Ga and 0.53 Ga, respectively). Note that some inferred LGTs could result from processes such as hybridization or allopolyploidization, and that these processes contribute dating information that can be treated in the same way as LGTs. On a wider scale, among eukaryotic groups, LGTs suggest that Amoebozoa (the outgroup, yellow in Fig. 4, estimated age of 0.85 Ga) diversified earlier than Opisthokonta and Apusozoa (the ingroup). This result indicates that LGTs could strongly reduce the uncertainty associated with the divergence of the major eukaryotic clades34.
Discussion
Our demonstration that clocks and transfers contain complementary and compatible dating signals casts the phylogenetic discord of LGTs in a new light, and calls for the development of new methods to combine these two types of dating information. Relaxed molecular clock models are fitted in a Bayesian framework, but current Markov Chain Monte Carlo proposal mechanisms can handle absolute, but not relative time constraints. Calibrating a molecular clock in a consistent probabilistic framework with both fossil-based and transfer-based time information will require modelling the effects of dependencies between separate parts of the tree, which current methods consider as independent. In the meantime, it is possible to partially take relative constraints into account in a typical relaxed clock analysis by two means. First, when fossil calibrations are available for some nodes, we can propagate their minimum age to all nodes constrained by transfers to be older, and, symmetrically, we can propagate their maximum age to all nodes constrained by transfers to be younger. Second, we can use rejection sampling; that is, discard posterior samples that fall below a threshold level of agreement with transfer-based constraints. These approaches, however, do not guarantee that all strongly supported relative constraints will be respected. To produce time-calibrated chronograms that respect all constraints (Fig. 4), we used a heuristic approach that indirectly estimates the age of nodes that are incompatible with constraints by interpolating between nodes whose ages do not violate the constraints.
The geological record of microbial life is sparse, and its interpretation is fraught with difficulty. Our results show that there is abundant information in extant genomes for dating the tree of life, and this information is waiting to be harvested to reconstruct genome evolution. This signal mostly contains information on the relative timing of diversification of groups that have exchanged genes through LGT, but we foresee several strategies for relating this relative timing to the broader history of life on Earth. First, gene transfers between bacteria and multicellular organisms that have left a trace in the fossil record will enable the propagation of absolute time calibrations to the microbial part of the tree of life35. Similarly, the signal of coevolution between hosts and their symbionts, such as in the gut microbiome of mammals36, could also be used to propagate absolute dating information from the host to the symbiont phylogeny. Finally, geochemistry can provide major constraints on early evolution37,38; for example, LGT events associated with ancestors of bacteria capable of oxygenic photosynthesis, that is, Oxyphotobacteria39, imply that the donor lineages must be older than the oxygenation of Earth’s atmosphere at approximately 2.3 Ga37,38. Phylogenetic models of genome evolution have the potential to turn the phylogenetic discord caused by gene transfer into an invaluable source of information for dating the tree of life.
Methods
We considered genome-scale datasets of homologous gene families from complete genomes in Cyanobacteria (40 genomes22), Archaea (60 genomes11) and Fungi (60 genomes23). For each gene family we used the species tree-aware probabilistic gene tree inference method ALE undated22,24 to sample evolutionary scenarios involving events of duplication, transfer and loss of genes conditional on a rooted species phylogeny and multiple sequence alignment of the family. The undated reconciliation method ignores tree branch lengths and does not impose any constraint on possible donor–recipient branch pairs aside from forbidding transfers to go from descendants to parents (Supplementary Fig. 9). For putative gene transfer events, we recorded the donor and recipient branches and used the frequency with which they occurred among the sampled scenarios to filter transfers and weight the relative age information they imply. Because the reference species tree is not dated, individual transfers can imply conflicting information about the relative age of speciation nodes (Supplementary Fig. 11). To extract a maximal subset of transfers consistent with each other, we used the newly developed optimization method MaxTiC25 (see also Supplementary Information). A maximal subset of consistent transfers specifies a time order of speciation events in the species tree. For instance, using MaxTiC on the 4,816 transfers that correspond to relative age constraints (Fig. 1b, Supplementary Figs. 8, 10) in the 5,322 gene families considered for Cyanobacteria, we identified a maximal subset of 3,322 (69%) transfers that are consistent (Supplementary Table 1). This maximal subset of transfers implies a time order of speciations that correlates with the distance between amino acid sequences of extant organisms (Spearman’s ρ = 0.741, P < 10−6; Fig. 1d, Supplementary Fig. 9). A similar correlation (Fig. 1c) can be observed if, following Zuckerkandl and Pauling1, we compare fossil dates and sequence divergence in mammals2 (10 time points, Pearson’s R2 = 0.664, P = 0.0025 and Spearman’s ρ = 0.83, P = 0.0056).
We used Phylobayes40 on a concatenate of nearly universal gene family alignments to sample chronograms (that is, dated trees) under four different uncalibrated molecular clock models41 (the strict molecular clock, the autocorrelated lognormal, the uncorrelated gamma, and the white-noise model). Chronograms were sampled using different calibration schemes described in the Supplementary Information and in the main text.
To estimate trees calibrated to geological time that obey transfer-based relative age constraints presented in Fig. 4, we followed a three-step approach. First, for each dataset we sampled 5,000 time orders compatible with LGT-based constraints obtained from MaxTiC. Second, for each dataset, we sampled chronograms calibrated to geological time with fossil calibrations using Phylobayes as described above (see also Supplementary Information and Supplementary Table 4) and assigned to each node of the phylogeny a direct age estimate corresponding to the median of the node ages in chronograms with top 5% agreement with LGT-based constraints obtained from MaxTiC (Supplementary Figs. 25–27). Finally, we calibrated each of the 5,000 time orders to geological time by removing conflicting node age estimates until we obtained a set of node ages compatible with the time order. Nodes left without node age estimates were assigned an indirect age corresponding to a random date distributed uniformly between the nearest existing dates such that the time order was obeyed. For each sampled time order, conflicting age estimates were removed in a fixed order corresponding to decreasing conflict calculated over all 5,000 sampled time orders, so that the ages that conflicted with the largest number of time orders were removed first.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
All data used in the study are available in the Supplementary Information or can be downloaded from the following website: ftp://pbil.univ-lyon1.fr/pub/datasets/davin2017/.
References
Zuckerkandl, E. & Pauling, L. in Horizons in Biochemistry (eds Kasha, M. & Pullman, B.) 189–225 (Academis Press, New York, 1962).
dos Reis, M. et al. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. Biol. Sci. 279, 3491–3500 (2012).
O’Leary, M. A. et al. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667 (2013).
Donoghue, P. C. J. & Benton, M. J. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Ecol. Evol. 22, 424–431 (2007).
Yang, Z. & Donoghue, P. C. J. Dating species divergences using rocks and clocks. Phil. Trans. R. Soc. B 371, 20150126 (2016).
Knoll, A. H. in Fundamentals of Geobiology (eds Knoll, A. H., Canfield, D. E. & Konhauser, K. O.) Ch. 16 (John Wiley, Chichester, 2012).
Knoll, A. H. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb. Perspect. Biol. 6, a0161211 (2014).
Doolittle, W. F. Phylogenetic classification and the Universal Tree. Science 284, 2124–2128 (1999).
Abby, S. S., Tannier, E., Gouy, M. & Daubin, V. Lateral gene transfer as a support for the tree of life. Proc. Natl Acad. Sci. USA 109, 4962–4967 (2012).
Szöllősi, G. J., Boussau, B., Abby, S. S., Tannier, E. & Daubin, V. Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations. Proc. Natl Acad. Sci. USA 109, 17513–17518 (2012).
Williams, T. A. et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl Acad. Sci. USA 114, E4602–E4611 (2017).
Huang, J. & Gogarten, J. P. Ancient gene transfer as a tool in phylogenetic reconstruction. Methods Mol. Biol. 532, 127–139 (2009).
Huang, J., Xu, Y. & Gogarten, J. P. The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Mol. Biol. Evol. 22, 2142–2146 (2005).
Wolfe, J. M. & Fournier, G. P. Tunneling through time: horizontal gene transfer constrains the timing of methanogen evolution. Preprint at https://www.biorxiv.org/content/early/2018/02/01/129494 (2017).
Maddison, W. P. Gene trees in species trees. Syst. Biol. 46, 523–536 (1997).
Szöllősi, G. J. & Daubin, V. Modeling gene family evolution and reconciling phylogenetic discord. Methods Mol. Biol. 856, 29–51 (2012).
Sjöstrand, J. et al. A Bayesian method for analyzing lateral gene transfer. Syst. Biol. 63, 409–420 (2014).
Szöllősi, G. J., Tannier, E., Daubin, V. & Boussau, B. The inference of gene trees with species trees. Syst. Biol. 64, e42–e62 (2015).
Daubin, V. & Szöllősi, G. J. Horizontal gene transfer and the history of life. Cold Spring Harb. Perspect. Biol. 8, a018036 (2016).
Gogarten, J. P., Murphey, R. D. & Olendzenski, L. Horizontal gene transfer: pitfalls and promises. Biol. Bull. 196, 359–362 (1999).
Szöllősi, G. J., Tannier, E., Lartillot, N. & Daubin, V. Lateral gene transfer from the dead. Syst. Biol. 62, 386–397 (2013).
Szöllősi, G. J., Davín, A. A., Tannier, E., Daubin, V. & Boussau, B. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Phil. Trans. R. Soc. B 370, 20140335 (2015).
Nagy, L. G. et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat. Commun. 5, 4471 (2014).
Szöllősi, G. J., Rosikiewicz, W., Boussau, B., Tannier, E. & Daubin, V. Efficient exploration of the space of reconciled gene trees. Syst. Biol. 62, 901–912 (2013).
Chauve, C. et al. MaxTiC: Fast ranking of a phylogenetic tree by maximum time consistency with lateral gene transfers. Preprint at https://www.biorxiv.org/content/early/2017/10/06/127548 (2017).
Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985).
Lepage, T., Bryant, D., Philippe, H. & Lartillot, N. A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24, 2669–2680 (2007).
Tomitani, A., Knoll, A. H., Cavanaugh, C. M. & Ohno, T. The evolutionary diversification of Cyanobacteria: molecular–phylogenetic and paleontological perspectives. Proc. Natl Acad. Sci. USA 103, 5442–5447 (2006).
Blank, C. E. & Sánchez-Baracaldo, P. Timing of morphological and ecological innovations in the cyanobacteria — a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23 (2010).
Shih, P. M., Hemp, J., Ward, L. M., Matzke, N. J. & Fischer, W. W. Crown group Oxyphotobacteria postdate the rise of oxygen. Geobiology 15, 19–29 (2017).
Dufresne, A., Garczarek, L. & Partensky, F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6, R14 (2005).
Ueno, Y., Yamada, K., Yoshida, N., Maruyama, S. & Isozaki, Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440, 516–519 (2006).
Spatafora, J. W. et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046 (2016).
Eme, L., Sharpe, S. C., Brown, M. W. & Roger, A. J. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 6, a016139 (2014).
Wybouw, N. et al. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife 3, e02365 (2014).
Groussin, M. et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 (2017).
Knoll, A. H., Bergmann, K. D. & Strauss, J. V. Life: the first two billion years. Phil. Trans. R. Soc. B 371, 20150493 (2016).
Wolfe, J. M. & Fournier, G. P. Tunneling through time: horizontal gene transfer constrains the timing of methanogen evolution. Preprint at https://www.biorxiv.org/content/early/2017/04/21/129494 (2017).
Soo, R. M., Hemp, J., Parks, D. H., Fischer, W. W. & Hugenholtz, P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 355, 1436–1440 (2017).
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004).
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009).
Benton, M. J. & Donoghue, P. C. J. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26–53 (2007).
dos Reis, M., Donoghue, P. C. J. & Yang, Z. Bayesian molecular clock dating of species divergences in the genomics era. Nat. Rev. Genet. 17, 71–80 (2016).
Acknowledgements
G.J.Sz. received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 714774. This project was supported by the French Agence Nationale de la Recherche through grant no. ANR-10-BINF-01–01 ‘Ancestrome’. Computations were performed using the Curie supercomputer thanks to PRACE project 2013081661 and the computing facilities of the CC LBBE/PRABI. T.A.W. is supported by a Royal Society University Research Fellowship. We thank N. Lartillot, T. Warnow, M. Paris, I. Derényi, L. Nagy and J. Miguel Blanca Postigo for discussions, comments on the manuscript and additional computing resources.
Author information
Authors and Affiliations
Contributions
E.T., B.B., V.D. and G.J.Sz. conceived the study. A.A.D., B.B., E.T. and G.J.Sz. developed the computational tools, T.A.W. contributed datasets, and A.A.D., E.T., B.B., T.A.W., V.D. and G.J.Sz. analysed the data, interpreted the results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figures 1–29, Supplementary Tables 1–4.
Supplementary Table 5
Constraints and their support in cyanobacteria.
Supplementary Table 6
Constraints and their support in archaea.
Supplementary Table 7
Constraints and their support in fungi.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Davín, A.A., Tannier, E., Williams, T.A. et al. Gene transfers can date the tree of life. Nat Ecol Evol 2, 904–909 (2018). https://doi.org/10.1038/s41559-018-0525-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-018-0525-3
This article is cited by
-
Dating Alphaproteobacteria evolution with eukaryotic fossils
Nature Communications (2021)
-
Evolution of the cytochrome bd oxygen reductase superfamily and the function of CydAA’ in Archaea
The ISME Journal (2021)
-
Heterothallism and potential hybridization events inferred for twenty-two yellow morel species
IMA Fungus (2020)
-
Isolation of an archaeon at the prokaryote–eukaryote interface
Nature (2020)