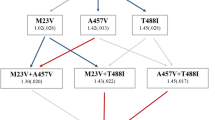
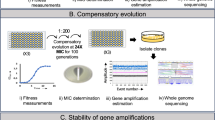
Abstract
Horizontal gene transfer mediated by broad-host-range plasmids is an important mechanism of antibiotic resistance spread. While not all bacteria maintain plasmids equally well, plasmid persistence can improve over time, yet no general evolutionary mechanisms have emerged. Our goal was to identify these mechanisms and to assess if adaptation to one plasmid affects the permissiveness to others. We experimentally evolved Pseudomonas sp. H2 containing multidrug resistance plasmid RP4, determined plasmid persistence and cost using a joint experimental–modelling approach, resequenced evolved clones, and reconstructed key mutations. Plasmid persistence improved in fewer than 600 generations because the fitness cost turned into a benefit. Improved retention of naive plasmids indicated that the host evolved towards increased plasmid permissiveness. Key chromosomal mutations affected two accessory helicases and the RNA polymerase β-subunit. Our and other findings suggest that poor plasmid persistence can be caused by a high cost involving helicase–plasmid interactions that can be rapidly ameliorated.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gogarten, J. P. & Townsend, J. P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679–687 (2005).
Schwarz, S. & Johnson, A. P. Transferable resistance to colistin: a new but old threat. J. Antimcrob. Chemother. 71, 2066–2070 (2016).
Baker, K. S. et al. The Murray collection of Enterobacteriacae: a unique resource. Genome Med. 7, 97–114 (2015).
Thomas, C. M. The Horizontal Gene Pool — Bacterial Plasmids and Gene Spread (Harwood Academic, Amsterdam, 2000).
Datta, N. & Hughes, V. M. Plasmids of the same Inc groups in enterobacteria before and after the medical use of antibiotics. Nature 306, 616–617 (1983).
Popowska, M. & Krawczyk-Balska, A. Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4, 1–8 (2013).
De Gelder, L., Ponciano, J. M., Joyce, P. & Top, E. M. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153, 452–463 (2007).
Ebersbach, G. & Gerdes, K. Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453–479 (2005).
Stewart, F. M. & Levin, B. R. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87, 209–228 (1977).
Harrison, E., Guymer, D., Spiers, A. J., Paterson, S. & Brockhurst, M. A. Parallel compensatory evolution stabilizes plasmids across the parasitism–mutualism continuum. Curr. Biol. 25, 2034–2039 (2015).
San Millan, A., Toll-Riera, M., Qi, Q. & MacLean, R. C. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 6, 6845 (2015).
Shintani, M. et al. Response of the Pseudomonas host chromosomal transcriptome to carriage of the IncP-7 plasmid pCAR1. Environ. Microbiol. 12, 1413–1426 (2010).
De Gelder, L., Williams, J. J., Ponciano, J. M., Sota, M. & Top, E. M. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178, 2179–2190 (2008).
Sota, M. et al. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 4, 1568–1580 (2010).
Bouma, J. E. & Lenski, R. E. Evolution of a bacteria/plasmid association. Nature 335, 351–352 (1988).
San Millan, A. et al. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208 (2014).
Dahlberg, C. & Chao, L. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165, 1641–1649 (2003).
Loftie-Eaton, W. et al. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol. Biol. Evol. 33, 885–897 (2016).
Maestro, B., Sanz, J. M., Díaz-Orejas, R. & Fernández-Tresguerres, E. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J. Bacteriol. 185, 1367–1375 (2003).
Yano, H. et al. Evolved plasmid–host interactions reduce plasmid interference cost. Mol. Microbiol. 101, 743–756 (2016).
Heuer, H., Fox, R. E. & Top, E. M. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 59, 738–748 (2007).
Pansegrau, W. et al. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and analysis. J. Mol. Biol. 239, 626–663 (1994).
Saunders, J. R. & Grinsted, J. Properties of RP4, an R factor which originated in Pseudomonas aeruginosa S8. J. Bacteriol. 112, 690–696 (1972).
Baquero, F., Coque, T. M. & de la Cruz, F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob. Agents Chemother. 55, 3649–3660 (2001).
Ponciano, J. M., De Gelder, L., Top, E. M. & Joyce, P. The population biology of bacterial plasmids: a hidden Markov model approach. Genetics 176, 957–968 (2007).
Lenski, R. E., Simpson, S. C. & Nguyen, T. T. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176, 3140–3147 (1994).
Heuer, H., Ebers, J., Weinert, N. & Smalla, K. Variation in permissiveness for broad-host-range plasmids among genetically indistinguishable isolates of Dickeya sp. from a small field plot. FEMS Microbiol. Ecol. 73, 190–196 (2010).
Klümper, U. et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945 (2014).
Kelley, A. L., Mezulis, S., Tayes, M. C., Wass, N. M. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Yachdav, G. et al. PredictProtein - an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, 337–343 (2014).
Byrd, A. K. & Raney, K. D. Superfamily 2 helicases. Front. Biosci. 17, 2070–2088 (2013).
Merrikh, H., Zhang., Y., Grossman, A. D. & Wang, J. D. Replication–transcription conflicts in bacteria. Nat. Rev. Microbiol. 10, 449–458 (2013).
Merrikh, C. N., Brewer, B. J. & Merrikh, H. The B. subtilis accessory helicase PcrA facilitates DNA replication through transcription units. PLoS Genet. 11, e1005289 (2015).
Fairman-Williams, M. E., Guenther, U. & Jankowsky, E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010).
Baharoglu, Z., Bikard, D. & Mazel, D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6, e1001165 (2010).
Ingmer, H., Miller, C. & Cohen, S. N. The RepA protein of plasmid pSC101 controls Escherichia coli cell division through the SOS response. Mol. Microbiol. 42, 519–526 (2001).
Boubakri, H., de Septenville, A. L., Viguera, E. & Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcriptional units in vivo. EMBO J. 29, 145–157 (2010).
Epshtein, V. et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505, 372–377 (2014).
Guy, C. P. et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36, 654–666 (2009).
Borukhou, S. & Nudler, E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 6, 93–100 (2003).
Kuznedelov, K. et al. A role for interaction of the RNA polymerase flap domain with σ subunit in promoter recognition. Science 295, 855–857 (2002).
Brandis, G., Wrande, M., Liljas, L. & Hughes, D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 85, 142–151 (2012).
Reynolds, M. G. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156, 1471–1481 (2000).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual (Cold Spring Harbour Laboratory Press, New York, 2001).
Joyce, P. et al. Modeling the impact of periodic bottlenecks, unidirectional mutation, and observational error in experimental evolution. J. Math. Biol. 50, 645–662 (2005).
Simonsen, L., Gordon, D. M., Stewart, F. M. & Levin, B. R. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136, 2319–2325 (1990).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Krzywinski, M. I. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Hmelo, L. R. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841 (2015).
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases grant R01 AI084918 of the National Institutes of Health (NIH). The genome resequencing was done by the IBEST Genomics Research Core and made possible thanks to an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) of the NIH under grant number P30 GM103324. K.B. was in part supported by a University of Idaho Department of Biological Sciences undergraduate research grant and by an NIGMS INBRE award, grant number P20 GM103408. H.Q. was supported by a National Science Foundation REU Site award, 1460696. We thank the laboratory of C. Marx for providing us with vector pPS04 and we thank H. Merrikh for useful suggestions.
Author information
Authors and Affiliations
Contributions
W.L. and E.M.T conceived the project and wrote the manuscript. W.L., K.B., H.Q., K.D., M.K.T. and J.M. performed the experiments. W.L. and J.M.P. performed the statistical analyses. W.L. and S.H. performed the bioinformatic analysis. H.M. facilitated part of the work and helped with data interpretation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Methods, Supplementary Figures 1–9 and Supplementary Tables 1–4, 6–8 and 10
Supplementary Data 1
Supplementary Tables 5 and 9
Rights and permissions
About this article
Cite this article
Loftie-Eaton, W., Bashford, K., Quinn, H. et al. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat Ecol Evol 1, 1354–1363 (2017). https://doi.org/10.1038/s41559-017-0243-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0243-2
This article is cited by
-
Plasmid-mediated phenotypic noise leads to transient antibiotic resistance in bacteria
Nature Communications (2024)
-
Horizontal gene transfer facilitates the molecular reverse-evolution of antibiotic sensitivity in experimental populations of H. pylori
Nature Ecology & Evolution (2024)
-
Oxytetracycline and heavy metals promote the migration of resistance genes in the intestinal microbiome by plasmid transfer
The ISME Journal (2023)
-
Regulatory fine-tuning of mcr-1 increases bacterial fitness and stabilises antibiotic resistance in agricultural settings
The ISME Journal (2023)
-
Bacterial volatile organic compounds attenuate pathogen virulence via evolutionary trade-offs
The ISME Journal (2023)