Abstract
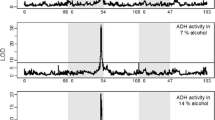
Identifying the genetic basis for adaptive differences between species requires explicit tests of historical hypotheses concerning the effects of past changes in gene sequence on molecular function, organismal phenotype and fitness. We address this challenge by combining ancestral protein reconstruction with biochemical experiments and physiological analysis of transgenic animals that carry ancestral genes. We tested a widely held hypothesis of molecular adaptation—that changes in the alcohol dehydrogenase protein (ADH) along the lineage leading to Drosophila melanogaster increased the catalytic activity of the enzyme and thereby contributed to the ethanol tolerance and adaptation of the species to its ethanol-rich ecological niche. Our experiments strongly refute the predictions of the adaptive ADH hypothesis and caution against accepting intuitively appealing accounts of historical molecular adaptation that are based on correlative evidence. The experimental strategy we employed can be used to decisively test other adaptive hypotheses and the claims they entail about past biological causality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sabeti, P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007).
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015).
Liu, S. et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014).
Shen, Y.-Y. et al. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl Acad. Sci. USA 107, 8666–8671 (2010).
Li, J. et al. Joint analysis of demography and selection in population genetics: where do we stand and where could we go? Mol. Ecol. 21, 28–44 (2012).
Storz, J. F. & Wheat, C. W. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution 64, 2489–2509 (2010).
Barrett, R. D. H. & Hoekstra, H. E. Molecular spandrels: tests of adaptation at the genetic level. Nat. Rev. Genet. 12, 767–780 (2011).
Lewontin, R. C. Twenty-five years ago in genetics: electrophoresis in the development of evolutionary genetics: milestone or millstone? Genetics 128, 657 (1991).
Lewontin, R. C. The Genetic Basis of Evolutionary Change (Columbia Univ. Press, 1974).
Feder, M. E. & Watt, W. B. in Genes in Ecology (eds Berry, R. J. et al. ) 365–392 (Blackwell Scientific, 1992).
Storz, J. F. et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl Acad. Sci. USA 106, 14450–14455 (2009).
Prasad, K. V. S. K. et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337, 1081–1084 (2012).
Tishkoff, S. A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007).
Linnen, C. R. et al. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339, 1312–1316 (2013).
Chan, Y. F. et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010).
Cheviron, Z. A. et al. Integrating evolutionary and functional tests of adaptive hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis . Mol. Biol. Evol. 31, 2948–2962 (2014).
Harms, M. J. & Thornton, J. W. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 20, 360–366 (2010).
Heinstra, P. W., Thörig, G. E., Scharloo, W., Drenth, W. & Nolte, R. J. Kinetics and thermodynamics of ethanol oxidation catalyzed by genetic variants of the alcohol dehydrogenase from Drosophila melanogaster and D. simulans . Biochim. Biophys. Acta 967, 224–233 (1988).
Freeman, S. & Herron, J. C. Evolutionary Analysis (Pearson Prentice Hall, 2007).
Johnson, N. A. Darwinian Detectives: Revealing the Natural History of Genes and Genomes (Oxford Univ. Press, 2007).
Kreitman, M., Shorrocks, B. & Dytham, C. in Genes in Ecology (eds Berry, R. J. et al. ) 281–312 (Blackwell Scientific, 1992).
Fox, C. W. & Wolf, J. B. Evolutionary Genetics: Concepts and Case Studies (Oxford Univ. Press, 2006).
Ruse, M. Darwin and Design: Does Evolution Have a Purpose? (Harvard Univ. Press, 2003).
Heinstra, P. W. H. Evolutionary genetics of the Drosophila alcohol dehydrogenase gene-enzyme system. Genetica 92, 1–22 (1993).
McKenzie, J. A. & Parsons, P. A. Alcohol tolerance: an ecological parameter in the relative success of Drosophila melanogaster and Drosophila simulans . Oecologia 10, 373–388 (1972).
McDonald, J. F. & Avise, J. C. Evidence for the adaptive significance of enzyme activity levels: interspecific variation in α-GPDH and ADH in Drosophila . Biochem. Genet. 14, 347–355 (1976).
Heinstra, P. W. H., Scharloo, W. & Thorig, G. E. W. Physiological significance of the alcohol dehydrogenase polymorphism in larvae of Drosophila . Genetics 117, 75–84 (1987).
McDonald, J. H. & Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila . Nature 351, 652–654 (1991).
Hartl, D. L. & Clark, A. G. Principles of Population Genetics 4th edn (Sinauer Associates, 2007).
Charlesworth, B. & Charlesworth, D. Population genetics from 1966 to 2016. Heredity (2016).
Dickinson, W. J., Rowan, R. G. & Brennan, M. D. Regulatory gene evolution: adaptive differences in expression of alcohol dehydrogenase in Drosophila melanogaster and Drosophila simulans . Heredity 52, 215–225 (1984).
Laurie, C. C., Heath, E. M., Jacobson, J. W. & Thomson, M. S. Genetic basis of the difference in alcohol dehydrogenase expression between Drosophila melanogaster and Drosophila simulans . Proc. Natl Acad. Sci. USA 87, 9674–9678 (1990).
Thomson, M. S., Jacobson, J. W. & Laurie, C. C. Comparison of alcohol dehydrogenase expression in Drosophila melanogaster and Drosophila simulans . Mol. Biol. Evol. 8, 31–48 (1991).
Montooth, K. L., Siebenthall, K. T. & Clark, A. G. Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster . J. Exp. Biol. 209, 3837–3850 (2006).
Kaun, K. R., Devineni, A. V. & Heberlein, U. Drosophila melanogaster as a model to study drug addiction. Hum. Genet. 131, 959–975 (2012).
Fry, J. D. Mechanisms of naturally evolved ethanol resistance in Drosophila melanogaster . J. Exp. Biol. 217, 3996–4003 (2014).
Sezgin, E. et al. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster . Genetics 168, 923–931 (2004).
Umina, P. A., Weeks, A. R., Kearney, M. R., McKechnie, S. W. & Hoffmann, A. A. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693 (2005).
Laurie, C. C. & Stam, L. F. The effect of an intronic polymorphism on alcohol dehydrogenase expression in Drosophila melanogaster . Genetics 138, 379–385 (1994).
Begun, D. J., Betancourt, A. J., Langley, C. H. & Stephan, W. Is the fast/slow allozyme variation at the Adh locus of Drosophila melanogaster an ancient balanced polymorphism? Mol. Biol. Evol. 16, 1816–1819 (1999).
Berry, A. & Kreitman, M. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134, 869–893 (1993).
Pool, J. E. et al. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8, e1003080 (2012).
Parsons, P. A. Ethanol utilization: threshold differences among six closely related species of Drosophila . Aust. J. Zool. 28, 535–541 (1980).
Stam, L. F. & Laurie, C. C. Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster . Genetics 144, 1559–1564 (1996).
Hudson, R. R., Kreitman, M. & Aguadé, M. A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 (1987).
Oakeshott, J. G. et al. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36, 86–96 (1982).
Yang, Z., Kumar, S. & Nei, M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650 (1995).
Loehlin, D. W. & Carroll, S. B. Expression of tandem gene duplicates is often greater than twofold. Proc. Natl Acad. Sci. USA 113, 5988–5992 (2016).
Eyre-Walker, A. Changing effective population size and the McDonald–Kreitman test. Genetics 162, 2017–2024 (2002).
Acknowledgements
We thank L. Picton, K. O’Brien, K. Gordon and members of the C. Meiklejohn and K. Montooth laboratories for technical assistance. We thank D. Matute for providing polymorphism data for D. yakuba. We thank M. Kreitman, members of the J. Thornton laboratory and D. Anderson for comments and suggestions that enriched the project. The project was supported by a National Science Foundation (NSF) grant (DEB-1501877; J.W.T./M.A.S.), an NSF graduate research fellowship (M.A.S.), National Institutes of Health (NIH) grant (R01-GM104397; J.W.T.), NSF CAREER Award (1505247; K.L.M.) and an NIH training grant (T32-GM007197; M.A.S.). D.W.L. was supported by a Howard Hughes Medical Institute postdoctoral fellowship from the Life Sciences Research Foundation and an investigatorship to S. B. Carroll from the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
M.A.S. and J.W.T. conceived the project. All authors participated in the experimental design. M.A.S. performed the phylogenetic and population genetic analyses. D.W.L. constructed the transgenic animals. M.A.S., D.W.L. and K.L.M. performed the functional experiments. All authors participated in data analysis and interpretation. M.A.S. and J.W.T. wrote the paper with contributions from D.W.L. and K.L.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1–4 and Supplementary Tables 1–4 (PDF 1005 kb)
Rights and permissions
About this article
Cite this article
Siddiq, M., Loehlin, D., Montooth, K. et al. Experimental test and refutation of a classic case of molecular adaptation in Drosophila melanogaster. Nat Ecol Evol 1, 0025 (2017). https://doi.org/10.1038/s41559-016-0025
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-016-0025
This article is cited by
-
The curious consistency of carbon biosignatures over billions of years of Earth-life coevolution
The ISME Journal (2021)
-
Different genetic basis for alcohol dehydrogenase activity and plasticity in a novel alcohol environment for Drosophila melanogaster
Heredity (2020)
-
Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs
Nature Structural & Molecular Biology (2020)
-
Genome editing retraces the evolution of toxin resistance in the monarch butterfly
Nature (2019)
-
Multinucleotide mutations cause false inferences of lineage-specific positive selection
Nature Ecology & Evolution (2018)