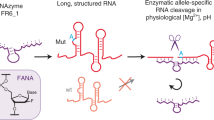
Abstract
Steric exclusion is a key element of enzyme substrate specificity, including in polymerases. Such substrate specificity restricts the enzymatic synthesis of 2′-modified nucleic acids, which are of interest in nucleic-acid-based drug development. Here we describe the discovery of a two-residue, nascent-strand, steric control ‘gate’ in an archaeal DNA polymerase. We show that engineering of the gate to reduce steric bulk in the context of a previously described RNA polymerase activity unlocks the synthesis of 2′-modified RNA oligomers, specifically the efficient synthesis of both defined and random-sequence 2′-O-methyl-RNA (2′OMe-RNA) and 2′-O-(2-methoxyethyl)-RNA (MOE-RNA) oligomers up to 750 nt. This enabled the discovery of RNA endonuclease catalysts entirely composed of 2′OMe-RNA (2′OMezymes) for the allele-specific cleavage of oncogenic KRAS (G12D) and β-catenin CTNNB1 (S33Y) mRNAs, and the elaboration of mixed 2′OMe-/MOE-RNA aptamers with high affinity for vascular endothelial growth factor. Our results open up these 2′-modified RNAs—used in several approved nucleic acid therapeutics—for enzymatic synthesis and a wider exploration in directed evolution and nanotechnology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information), except raw sequencing reads, which are available in the NCBI SRA repository, BioProject ID PRJNA847930. Source data are provided with this paper.
Code availability
Fidelity analysis of sequencing data for 2′OMe-RNA synthesis by individual polymerases was performed using the Burrows-Wheeler Aligner (BWA-0.7.17), Samtools and custom scripts, which can be found at GitHub: https://github.com/holliger-lab/fidelity-analysis.
References
Taylor, A. I., Houlihan, G. & Holliger, P. Beyond DNA and RNA: the expanding toolbox of synthetic genetics. Cold Spring Harb. Perspect. Biol. 11, a032490 (2019).
Freund, N., Furst, M. & Holliger, P. New chemistries and enzymes for synthetic genetics. Curr. Opin. Biotechnol. 74, 129–136 (2021).
Wan, W. B. & Seth, P. P. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 59, 9645–9667 (2016).
Dimitrova, D. G., Teysset, L. & Carre, C. RNA 2′-O-methylation (Nm) modification in human diseases. Genes 10, 117 (2019).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Zust, R. et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 (2011).
Aartsma-Rus, A. & Corey, D. R. The 10th oligonucleotide therapy approved: golodirsen for Duchenne muscular dystrophy. Nucleic Acid Ther. 30, 67–70 (2020).
Ruckman, J. et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273, 20556–20567 (1998).
Chelliserrykattil, J. & Ellington, A. D. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat. Biotechnol. 22, 1155–1160 (2004).
Ibach, J. et al. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2′-O-methyl-modified RNA. J. Biotechnol. 167, 287–295 (2013).
Meyer, A. J. et al. Transcription yield of fully 2′-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res. 43, 7480–7488 (2015).
Burmeister, P. E. et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 12, 25–33 (2005).
Chen, T. et al. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2′-modified DNA. Nat. Chem. 8, 556–562 (2016).
Liu, Z., Chen, T. & Romesberg, F. E. Evolved polymerases facilitate selection of fully 2′-OMe-modified aptamers. Chem. Sci. 8, 8179–8182 (2017).
Hoshino, H., Kasahara, Y., Kuwahara, M. & Obika, S. DNA polymerase variants with high processivity and accuracy for encoding and decoding locked nucleic acid sequences. J. Am. Chem. Soc. 142, 21530–21537 (2020).
Cozens, C. et al. Enzymatic synthesis of nucleic acids with defined regioisomeric 2′-5′ linkages. Angew. Chem. Int. Ed. Engl. 54, 15570–15573 (2015).
Cozens, C., Pinheiro, V. B., Vaisman, A., Woodgate, R. & Holliger, P. A short adaptive path from DNA to RNA polymerases. Proc. Natl Acad. Sci. USA 109, 8067–8072 (2012).
Kropp, H. M., Betz, K., Wirth, J., Diederichs, K. & Marx, A. Crystal structures of ternary complexes of archaeal B-family DNA polymerases. PLoS ONE 12, e0188005 (2017).
Perera, R. L. et al. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. eLife 2, e00482 (2013).
Kawai, G. et al. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry 31, 1040–1046 (1992).
Nishizaki, T., Iwai, S., Ohtsuka, E. & Nakamura, H. Solution structure of an RNA·2′-O-methylated RNA hybrid duplex containing an RNA·DNA hybrid segment at the center. Biochemistry 36, 2577–2585 (1997).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Houlihan, G. et al. Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity. Nat. Chem. 12, 683–690 (2020).
Taylor, A. I. & Holliger, P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 10, 1625–1642 (2015).
Egli, M. et al. Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2′-O-ribonucleic acid modifications. Biochemistry 44, 9045–9057 (2005).
Teplova, M. et al. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 6, 535–539 (1999).
Khatsenko, O. et al. Absorption of antisense oligonucleotides in rat intestine: effect of chemistry and length. Antisense Nucleic Acid Drug Dev. 10, 35–44 (2000).
Plevnik, M., Cevec, M. & Plavec, J. NMR structure of 2′-O-(2-methoxyethyl) modified and C5-methylated RNA dodecamer duplex. Biochimie 95, 2385–2391 (2013).
Martin, P. Stereoselektive Synthese von 2′-O-(2-Methoxyethyl)ribonucleosiden: Nachbargruppenbeteiligung der Methoxyethoxy-Gruppe bei der Ribosylierung von Heterocyclen. Helv. Chim. Acta 79, 1930–1938 (1996).
Martin, P. Ein neuer Zugang zu 2′-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotide. Helv. Chim. Acta 78, 486–504 (1995).
Gillerman, I. & Fischer, B. An improved one-pot synthesis of nucleoside 5′-triphosphate analogues. Nucleosides Nucleotides Nucleic Acids 29, 245–256 (2010).
Ludwig, J. A new route to nucleoside 5′-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung. 16, 131–133 (1981).
Freier, S. M. & Altmann, K. H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 25, 4429–4443 (1997).
Kool, E. T. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu. Rev. Biophys. Biomol. Struct. 30, 1–22 (2001).
Wu, E. Y. & Beese, L. S. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an ‘ajar’ intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 286, 19758–19767 (2011).
Wang, W., Wu, E. Y., Hellinga, H. W. & Beese, L. S. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J. Biol. Chem. 287, 28215–28226 (2012).
Chen, C. Y. DNA polymerases drive DNA sequencing-by-synthesis technologies: both past and present. Front. Microbiol. 5, 305 (2014).
Redrejo-Rodriguez, M. et al. Primer-independent DNA synthesis by a family B DNA polymerase from self-replicating mobile genetic elements. Cell Rep. 21, 1574–1587 (2017).
Blasco, M. A., Mendez, J., Lazaro, J. M., Blanco, L. & Salas, M. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J. Biol. Chem. 270, 2735–2740 (1995).
Kazlauskas, D., Krupovic, M., Guglielmini, J., Forterre, P. & Venclovas, C. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 48, 10142–10156 (2020).
Franklin, M. C., Wang, J. & Steitz, T. A. Structure of the replicating complex of a pol α family DNA polymerase. Cell 105, 657–667 (2001).
Rudinger, N. Z., Kranaster, R. & Marx, A. Hydrophobic amino acid and single-atom substitutions increase DNA polymerase selectivity. Chem. Biol. 14, 185–194 (2007).
Gardner, A. F. & Jack, W. E. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 27, 2545–2553 (1999).
Bartel, D. P. & Szostak, J. W. Isolation of new ribozymes from a large pool of random sequences. Science 261, 1411–1418 (1993).
Wilson, D. S. & Szostak, J. W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68, 611–647 (1999).
Mutschler H, et al. Random-sequence genetic oligomer pools display an innate potential for ligation and recombination. eLife 7, e43022 (2018).
Fedor, M. J. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297, 269–291 (2000).
Skerra, A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151, 131–135 (1994).
Taylor, A. I. et al. Catalysts from synthetic genetic polymers. Nature 518, 427–430 (2015).
Potty, A. S. et al. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers 91, 145–156 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
This work was supported by a PhD fellowship from Boehringer Ingelheim Fonds (N.F.), the Medical Research Council (MRC) programme (MC_U105178804; A.I.T., S.-Y.P.-C., P. Holliger), a research collaboration between AstraZeneca UK and the MRC (MRC–AstraZeneca Blue Sky Grant; N.S., S.A.-F.), FWO (Flanders Research Foundation) Fund of Scientific Research and the Rega Institute, KU Leuven (M.A., P. Herdewijn), and by National Institute of Health grants R35-GM128562 (B.D.F.) and K99-ES031148 (A.M.W.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
N.F., S.A.-F., A.I.T. and P. Holliger conceived and designed experiments. N.F. performed polymerase studies. N.F. and S.A.-F. performed polymerase design and engineering. N.F., M.A. and P. Herdewijn synthesized MOE-nucleotides. N.F. and A.I.T. completed SPR measurements. A.I.T. performed 2′OMezyme selections and characterization. N.S. performed polymerase fidelity measurements. S.-Y.P.-C. performed MS analysis. A.M.W. and B.D.F performed and analysed steady-state kinetic measurements. All authors analysed data, discussed results and co-wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
UK Research and Innovation has filed a UK patent priority application on behalf of the inventors N.F., S.A.-F. and P. Holliger on the 2M/3M polymerase on 25 May 2022 (application number 2207699.6).
Peer review
Peer review information
Nature Chemistry thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Full materials and methods, Supplementary Figs 1–17, Tables 1–7, references.
Supplementary Data 1
Statistical source data for graphs in Supplementary Fig. 2.
Supplementary Data 2
Statistical source data: raw Biacore SPR data that were fitted and plotted in Supplementary Fig. 9a.
Supplementary Data 3
Statistical source data: raw Biacore SPR data that were fitted and plotted in Supplementary Fig. 9b.
Source data
Source Data Fig. 1
Unprocessed gels for Fig. 1.
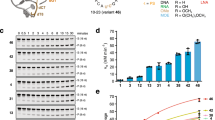
Source Data Fig. 2
Unprocessed gels for Fig. 2.
Source Data Fig. 2
Statistical source data for graphs in Fig. 2.
Source Data Fig. 3
Unprocessed gels for Fig. 3.
Source Data Fig. 4a
Statistical source data: raw Biacore SPR data that were fitted and plotted in Fig. 4a.
Source Data Fig. 4b
Statistical source data: raw Biacore SPR data that were fitted and plotted in Fig. 4b.
Source Data Fig. 4c
Statistical source data: raw Biacore SPR data that were fitted and plotted in Fig. 4c.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Freund, N., Taylor, A.I., Arangundy-Franklin, S. et al. A two-residue nascent-strand steric gate controls synthesis of 2′-O-methyl- and 2′-O-(2-methoxyethyl)-RNA. Nat. Chem. 15, 91–100 (2023). https://doi.org/10.1038/s41557-022-01050-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-022-01050-8