Abstract
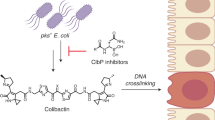
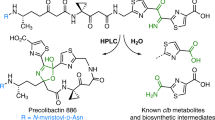
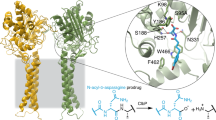
Colibactin is an assumed human gut bacterial genotoxin, whose biosynthesis is linked to the clb genomic island that has a widespread distribution in pathogenic and commensal human enterobacteria. Colibactin-producing gut microbes promote colon tumour formation and enhance the progression of colorectal cancer via cellular senescence and death induced by DNA double-strand breaks (DSBs); however, the chemical basis that contributes to the pathogenesis at the molecular level has not been fully characterized. Here, we report the discovery of colibactin-645, a macrocyclic colibactin metabolite that recapitulates the previously assumed genotoxicity and cytotoxicity. Colibactin-645 shows strong DNA DSB activity in vitro and in human cell cultures via a unique copper-mediated oxidative mechanism. We also delineate a complete biosynthetic model for colibactin-645, which highlights a unique fate of the aminomalonate-building monomer in forming the C-terminal 5-hydroxy-4-oxazolecarboxylic acid moiety through the activities of both the polyketide synthase ClbO and the amidase ClbL. This work thus provides a molecular basis for colibactin’s DNA DSB activity and facilitates further mechanistic study of colibactin-related colorectal cancer incidence and prevention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all the data supporting the findings of this study are available within the paper and the Supplementary Information, and/or from the corresponding authors upon reasonable request.
References
Nicholson, J. K. et al. Host–gut microbiota metabolic interactions. Science 336, 1262–1267 (2012).
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Sharon, G. et al. Specialized metabolites from the microbiome in health and disease. Cell Metab. 20, 719–730 (2014).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014).
Donia, M. S. & Fischbach, M. A. Small molecules from the human microbiota. Science 349, 1254766 (2015).
Bode, H. B. The microbes inside us and the race for colibactin. Angew. Chem. Int. Ed. 54, 10408–10411 (2015).
Balskus, E. P. Colibactin: understanding an elusive gut bacterial genotoxin. Nat. Prod. Rep. 32, 1534–1540 (2015).
Faïs, T., Delmas, J., Barnich, N., Bonnet, R. & Dalmasso, G. Colibactin: more than a new bacterial toxin. Toxins 10, e151 (2018).
Nougayrède, J. P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Cuevas-Ramos, G. et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl Acad. Sci. USA 107, 11537–11542 (2010).
Secher, T., Samba-Louaka, A., Oswald, E. & Nougayrède, J. P. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLoS ONE 8, e77157 (2013).
Cougnoux, A. et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63, 1932–1942 (2014).
Payros, D. et al. Maternally acquired genotoxic Escherichia coli alters offspring’s intestinal homeostasis. Gut Microbes 5, 313–325 (2014).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Tomkovich, S. et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 77, 2620–2632 (2017).
Buc, E. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 8, e56964 (2013).
Bondarev, V. et al. The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 15, 2095–2113 (2013).
Engel, P., Vizcaino, M. I. & Crawford, J. M. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl. Environ. Microbiol. 81, 1502–1512 (2015).
Brotherton, C. A. & Balskus, E. P. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J. Am. Chem. Soc. 135, 3359–3362 (2013).
Bian, X. In vivo evidence for a prodrug activation mechanism during colibactin maturation. ChemBioChem 14, 1194–1197 (2013).
Vizcaino, M. I., Engel, P., Trautman, E. & Crawford, J. M. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J. Am. Chem. Soc. 136, 9244–9247 (2014).
Brotherton, C. A., Wilson, M., Byrd, G. & Balskus, E. P. Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. Org. Lett. 17, 1545–1548 (2015).
Bian, X., Plaza, A., Zhang, Y. & Müller, R. Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem. Sci. 6, 3154–3160 (2015).
Vizcaino, M. I. & Crawford, J. M. The colibactin warhead crosslinks DNA. Nat. Chem. 7, 411–417 (2015).
Li, Z.-R. et al. Critical intermediates reveal new biosynthetic events in the enigmatic colibactin pathway. ChemBioChem 16, 1715–1719 (2015).
Brachmann, A. O. et al. Colibactin biosynthesis and biological activity depends on the rare aminomalonyl polyketide precursor. Chem. Commun. 51, 13138–13141 (2015).
Zha, L., Wilson, M. R., Brotherton, C. A. & Balskus, E. P. Characterization of polyketide synthase machinery from the pks island facilitates isolation of a candidate precolibactin. ACS Chem. Biol. 11, 1287–1295 (2016).
Li, Z.-R. et al. Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nat. Chem. Biol. 12, 773–775 (2016).
Zha, L. et al. Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nat. Chem. Biol. 13, 1063–1065 (2017).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001).
Guntaka, N. S., Healy, A. R., Crawford, J. M., Herzon, S. B. & Bruner, S. D. Structure and functional analysis of ClbQ, an unusual intermediate-releasing thioesterase from the colibactin biosynthetic pathway. ACS Chem. Biol. 12, 2598–2608 (2017).
Bossuet-Greif, N. et al. Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 99, 897–908 (2016).
Tripathi, P. et al. ClbS is a cyclopropane hydrolase that confers colibactin resistance. J. Am. Chem. Soc. 139, 17719–17722 (2017).
Colis, L. C. et al. The cytotoxicity of (–)-lomaiviticin A arises from induction of double-strand breaks in DNA. Nat. Chem. 6, 504–510 (2014).
Melvin, M. S. et al. Double-strand DNA cleavage by copper·prodigiosin. J. Am. Chem. Soc. 122, 6333–6334 (2000).
Povirk, L. F., Wübker, W., Köhnlein, W. & Hutchinson, F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 4, 3573–3580 (1977).
Stubbe, J. A. & Kozarich, J. W. Mechanisms of bleomycin-induced DNA degradation. Chem. Rev. 87, 1107–1136 (1987).
Chen, J. & Stubbe, J. Bleomycins: towards better therapeutics. Nat. Rev. Cancer 5, 102–112 (2005).
Pitie, M. & Pratviel, G. Activation of DNA carbon–hydrogen bonds by metal complexes. Chem. Rev. 110, 1018–1059 (2010).
Chaturvedi, K. S., Hung, C. S., Crowley, J. R., Stapleton, A. E. & Henderson, J. P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736 (2012).
Humphreys, K. J., Johnson, A. E., Karlin, K. D. & Rokita, S. E. Oxidative strand scission of nucleic acids by a multinuclear copper(ii) complex. J. Biol. Inorg. Chem. 7, 835–842 (2002).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Schultz, L. B., Chehab, N. H., Malikzay, A. & Halazonetis, T. D. p53 binding protein 1 (53bp1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151, 1381–1390 (2000).
Collins, A. The comet assay for DNA damage and repair. Mol. Biotechnol. 26, 249–261 (2004).
Burger, R. M., Peisach, J. & Horwitz, S. B. Activated bleomycin: a transient complex of drug, iron, and oxygen that degrades DNA. J. Biol. Chem. 256, 11636–11644 (1981).
Chan, Y. A. et al. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl. Acad. Sci. USA 103, 14349–14354 (2006).
Holmes, T. C. et al. Molecular insights into the biosynthesis of guadinomine: a type III secretion system inhibitor. J. Am. Chem. Soc. 134, 17797–17806 (2012).
Jiang, Y. et al. The reactivity of an unusual amidase may explain colibactin’s DNA cross-linking activity. Preprint at bioRxiv https://doi.org/10.1101/567248 (2019).
Xue, M. et al. Structure elucidation of colibactin. Preprint at bioRxiv https://doi.org/10.1101/574053 (2019).
Stern, B. R. et al. Copper and human health: biochemistry, genetics, and strategies for modeling dose–response relationships. J. Toxicol. Env. Heal. B 10, 157–222 (2007).
Borah, S., Melvin, M. S., Lindquist, N. & Manderville, R. A. Copper-mediated nuclease activity of a tambjamine alkaloid. J. Am. Chem. Soc. 120, 4557–4562 (1998).
Pommier, Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 8, 82–95 (2013).
Woo, C. M., Li, Z., Paulson, E. K. & Herzon, S. B. Structural basis for DNA cleavage by the potent antiproliferative agent (–)-lomaiviticin A. Proc. Natl Acad. Sci. USA 113, 2851–2856 (2016).
Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019).
Bossuet-Greif, N. et al. The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9, e02393-17 (2018).
Olier, M. et al. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3, 501–509 (2012).
Pérez-Berezo, T. et al. Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle 1917. Nat. Commun. 8, 1314 (2017).
Bleich, R. M. & Arthur, J. C. Revealing a microbial carcinogen. Science 363, 689–690 (2019).
Acknowledgements
This work was supported by an NSFC grant (no. 41576140), a China Ocean Mineral Resources Research and Development Association grant (COMRRDA17SC01) a grant from National Key R&D Programmes of China and a grant from the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) to P.-Y.Q., grants from the National Institutes of Health (DP2AT009148), the Alfred P. Sloan Foundation and the Chan Zuckerberg Biohub Investigator Program to W.Z. and a grant from the National Institutes of Health (R01-GM85770) to B.S.M. We thank D. Lin and L. Feng for NMR measurements, Y. K. Tam and N. Harris for assistance with mass spectrometry experiments, Y. Huang and S. Jia for helpful discussions and A. Cheung for manuscript proofreading. Z.-R.L. acknowledges support from the International House at UC Berkeley, Chevron Corporation and UC Berkeley as the fellow of Chevron-Xenel PhD Gateway Fellowship. S.M.K.M. acknowledges the NSERC-PDF fellowship.
Author information
Authors and Affiliations
Contributions
Z.-R.L. performed the experiments, J.Y.H.L. conducted some of the fermentation work, Z.-R.L., J.L., W.C. and S.M.K.M. analysed NMR data, Z.-R.L. and W.-P.Z. performed the gene analysis, Z.-R.L., W.Z. and P.-Y.Q. designed the study and wrote the manuscript, with input from J.L. and B.S.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Materials and Methods, Supplementary text, Supplementary Figs. 1–27, Supplementary Tables 1–7 and Supplementary references 1–28.
Rights and permissions
About this article
Cite this article
Li, ZR., Li, J., Cai, W. et al. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat. Chem. 11, 880–889 (2019). https://doi.org/10.1038/s41557-019-0317-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-019-0317-7
This article is cited by
-
Intratumoral microorganisms in tumors of the digestive system
Cell Communication and Signaling (2024)
-
Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy
Signal Transduction and Targeted Therapy (2023)
-
Enabling programmable dynamic DNA chemistry using small-molecule DNA binders
Nature Communications (2023)
-
Stool pattern is associated with not only the prevalence of tumorigenic bacteria isolated from fecal matter but also plasma and fecal fatty acids in healthy Japanese adults
BMC Microbiology (2021)
-
Reply to: Macrocyclic colibactins
Nature Chemistry (2020)