Abstract
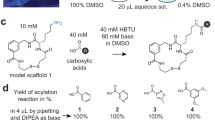
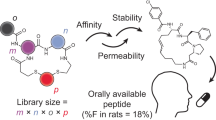
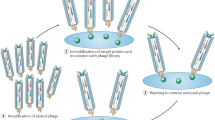
Successful screening campaigns depend on large and structurally diverse collections of compounds. In macrocycle screening, variation of the molecular scaffold is important for structural diversity, but so far it has been challenging to diversify this aspect in large combinatorial libraries. Here, we report the cyclization of peptides with two chemical bridges to provide rapid access to thousands of different macrocyclic scaffolds in libraries that are easy to synthesize, screen and decode. Application of this strategy to phage-encoded libraries allowed for the screening of an unprecedented structural diversity of macrocycles against plasma kallikrein, which is important in the swelling disorder hereditary angioedema. These libraries yielded inhibitors with remarkable binding properties (subnanomolar Ki, >1,000-fold selectivity) despite the small molecular mass (~1,200 Da). An interlaced bridge format characteristic of this strategy provided high proteolytic stability (t1/2 in plasma of >3 days), making double-bridged peptides potentially amenable to topical or oral delivery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Driggers, E. M., Hale, S. P., Lee, J. & Terrett, N. K. The exploration of macrocycles for drug discovery—an underexploited structural class. Nat. Rev. Drug Discov. 7, 608–624 (2008).
Levin, J. Macrocycles in Drug Discovery (Royal Society of Chemistry, Cambridge, 2014).
Villar, E. A. et al. How proteins bind macrocycles. Nat. Chem. Biol. 10, 723–731 (2014).
Giordanetto, F. & Kihlberg, J. Macrocyclic drugs and clinical candidates: what can medicinal chemists learn from their properties? J. Med. Chem. 57, 278–295 (2014).
Dow, M., Fisher, M., James, T., Marchetti, F. & Nelson, A. Towards the systematic exploration of chemical space. Org. Biomol. Chem. 10, 17–28 (2012).
Collins, S., Bartlett, S., Nie, F. L., Sore, H. F. & Spring, D. R. Diversity-oriented synthesis of macrocycle libraries for drug discovery and chemical biology. Synth. Stuttg. 48, 1457–1473 (2016).
Chen, S. & Heinis, C. in Biotherapeutics: Recent Developments using Chemical and Molecular Biology (eds Jones, L. & McKnight, A. J.) 241–262 (Royal Society of Chemistry, Cambridge, 2013).
Josephson, K., Ricardo, A. & Szostak, J. W. mRNA display: from basic principles to macrocycle drug discovery. Drug Discov. Today 19, 388–399 (2014).
Ladner, R. C., Sato, A. K., Gorzelany, J. & de Souza, M. Phage display-derived peptides as a therapeutic alternatives to antibodies. Drug Discov. Today 9, 525–529 (2004).
Obexer, R., Walport, L. J. & Suga, H. Exploring sequence space: harnessing chemical and biological diversity towards new peptide leads. Curr. Opin. Chem. Biol. 38, 52–61 (2017).
Heinis, C. & Winter, G. Encoded libraries of chemically modified peptides. Curr. Opin. Chem. Biol. 26, 89–98 (2015).
Ng, S., Jafari, M. R. & Derda, R. Bacteriophages and viruses as a support for organic synthesis and combinatorial chemistry. ACS Chem. Biol. 7, 123–138 (2012).
Bashiruddin, N. K., Nagano, M. & Suga, H. Synthesis of fused tricyclic peptides using a reprogrammed translation system and chemical modification. Bioorg. Chem. 61, 45–50 (2015).
Hacker, D. E., Hoinka, J., Iqbal, E. S., Przytycka, T. M. & Hartman, M. C. T. Highly constrained bicyclic scaffolds for the discovery of protease-stable peptides via mRNA display. ACS Chem. Biol. 12, 795–804 (2017).
Sako, Y., Morimoto, J., Murakami, H. & Suga, H. Ribosomal synthesis of bicyclic peptides via two orthogonal inter-side-chain reactions. J. Am. Chem. Soc. 130, 7232–7234 (2008).
Hayashi, Y., Morimoto, J. & Suga, H. In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem. Biol. 7, 607–613 (2012).
Rebollo, I. R., Angelini, A. & Heinis, C. Phage display libraries of differently sized bicyclic peptides. MedChemComm 4, 145–150 (2013).
Chen, S., Bertoldo, D., Angelini, A., Pojer, F. & Heinis, C. Peptide ligands stabilized by small molecules. Angew. Chem. Int. Ed. 53, 1602–1606 (2014).
Chua, K. et al. Small cyclic agonists of iron regulatory hormone hepcidin. Bioorg. Med. Chem. Lett. 25, 4961–4969 (2015).
Jo, H. et al. Development of α-helical calpain probes by mimicking a natural protein–protein interaction. J. Am. Chem. Soc. 134, 17704–17713 (2012).
Kowalczyk, R. et al. Synthesis and evaluation of disulfide bond mimetics of amylin-(1–8) as agents to treat osteoporosis. Bioorg. Med. Chem. Lett. 20, 2661–2668 (2012).
Bellotto, S., Chen, S., Rentero Rebollo, I., Wegner, H. A. & Heinis, C. Phage selection of photoswitchable peptide ligands. J. Am. Chem. Soc. 136, 5880–5883 (2014).
Jafari, M. R. et al. Discovery of light-responsive ligands through screening of a light-responsive genetically encoded library. ACS Chem. Biol. 9, 443–450 (2014).
Ng, S. & Derda, R. Phage-displayed macrocyclic glycopeptide libraries. Org. Biomol. Chem. 14, 5539–5545 (2016).
Schlippe, Y. V., Hartman, M. C., Josephson, K. & Szostak, J. W. In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors. J. Am. Chem. Soc. 134, 10469–10477 (2012).
Cicardi, M. et al. Ecallantide for the treatment of acute attacks in hereditary angioedema. New Engl. J. Med. 363, 523–531 (2010).
Plosker, G. L. Recombinant human C1 inhibitor (conestat alfa) in the treatment of angioedema attacks in hereditary angioedema. Biodrugs 26, 315–323 (2012).
Banerji, A. et al. Inhibiting plasma kallikrein for hereditary angioedema prophylaxis. New Engl. J. Med. 376, 717–728 (2017).
Smeenk, L. E. J., Dailly, N., Hiemstra, H., Maarseveen, J. H. & Timmerman, P. Synthesis of water-soluble scaffolds for peptide cyclization, labeling, and ligation. Org. Lett. 14, 1194–1197 (2012).
Timmerman, P., Beld, J., Puijk, W. C. & Meloen, R. H. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. ChemBioChem 6, 821–824 (2005).
Wang, Y. & Chou, D. H. C. A thiol-ene coupling approach to native peptide stapling and macrocyclization. Angew. Chem. Int. Ed. 54, 10931–10934 (2015).
Woolley, G. A. Photocontrolling peptide alpha helices. Acc. Chem. Res. 38, 486–493 (2005).
Assem, N., Ferreira, D. J., Wolan, D. W. & Dawson, P. E. Acetone-linked peptides: a convergent approach for peptide macrocyclization and labeling. Angew. Chem. Int. Ed. 54, 8665–8668 (2015).
Lee, M. T. W., Maruani, A. & Chudasama, V. The use of 3,6-pyridazinediones in organic synthesis and chemical biology. J. Chem. Res. 40, 1–9 (2016).
Smith, M. E. B. et al. Protein modification, bioconjugation, and disulfide bridging using bromomaleimides. J. Am. Chem. Soc. 132, 1960–1965 (2010).
Li, Z. et al. Structure-guided design of novel, potent, and selective macrocyclic plasma kallikrein inhibitors. ACS Med. Chem. Lett. 8, 185–190 (2017).
Tse, M. T. IL-17 antibodies gain momentum. Nat. Rev. Drug Discov. 12, 815–816 (2013).
Baeriswyl, V. et al. Bicyclic peptides with optimized ring size inhibit human plasma kallikrein and its orthologues while sparing paralogous proteases. ChemMedChem 7, 1173–1176 (2012).
Acknowledgements
This work was supported by the NCCR Chemical Biology of the Swiss National Science Foundation.
Author information
Authors and Affiliations
Contributions
S.S.K., C.V., X.-D.K. and C.H. conceived the strategy, designed experiments, analysed data and wrote the manuscript. S.S.K. established the chemical reactions. C.V. and X.-D.K. performed the phage selections. S.S.K., C.V. and X.-D.K. synthesized, purified and characterized peptides. A.Z. synthesized a linker reagent. K.D. contributed to the writing of the manuscript. S.S.K., C.V. and X.-D.K. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
C.H. is a scientific founder of Bicycle Therapeutics. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Materials and Methods, Tables 1–3, and Figures 1–13
Supplementary Slide Show
An animated slide show illustrating graphically the large scaffold diversity that can be generated
Supplementary Slide Show
An animated slide show illustrating graphically the large scaffold diversity that can be generated
Rights and permissions
About this article
Cite this article
Kale, S.S., Villequey, C., Kong, XD. et al. Cyclization of peptides with two chemical bridges affords large scaffold diversities. Nature Chem 10, 715–723 (2018). https://doi.org/10.1038/s41557-018-0042-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0042-7
This article is cited by
-
De novo development of small cyclic peptides that are orally bioavailable
Nature Chemical Biology (2024)
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)
-
An amide to thioamide substitution improves the permeability and bioavailability of macrocyclic peptides
Nature Communications (2023)
-
High-affinity peptides developed against calprotectin and their application as synthetic ligands in diagnostic assays
Nature Communications (2023)
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)