Abstract
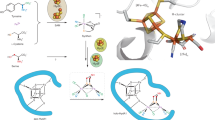
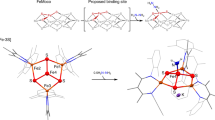
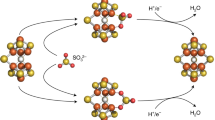
The M-cluster is the [(homocitrate)MoFe7S9C] active site of nitrogenase that is derived from an 8Fe core assembled viacoupling and rearrangement of two [Fe4S4] clusters concomitant with the insertion of an interstitial carbon and a ‘ninth sulfur’. Combining synthetic [Fe4S4] clusters with an assembly protein template, here we show that sulfite can give rise to the ninth sulfur that is incorporated in the catalytically important belt region of the cofactor after the radical S-adenosyl-l-methionine-dependent carbide insertion and the concurrent 8Fe-core rearrangement have already taken place. Based on the differential reactivity of the formed cluster species, we also propose a new [Fe8S8C] cluster intermediate, the L*-cluster, which is similar to the [Fe8S9C] L-cluster, but lacks the ninth sulfur from sulfite. This work provides a semi-synthetic tool for protein reconstitution that could be widely applicable for the functional analysis of other FeS systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burgess, B. K. & Lowe, D. J. Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012 (1996).
Rees, D. C. et al. Structural basis of biological nitrogen fixation. Phil. Trans. R. Soc. A 363, 971–984 (2005).
Chan, M. K., Kim, J. & Rees, D. C. The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 Å resolution structures. Science 260, 792–794 (1993).
Einsle, O. et al. Nitrogenase MoFe-protein at 1.16 Å resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700 (2002).
Spatzal, T. et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940 (2011).
Lancaster, K. M. et al. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron–molybdenum cofactor. Science 334, 974–977 (2011).
Wiig, J. A., Hu, Y. & Ribbe, M. W. NifEN-B complex of Azotobacter vinelandii is fully functional in nitrogenase FeMo cofactor assembly. Proc. Natl Acad. Sci. USA 108, 8623–8627 (2011).
Wiig, J. A., Hu, Y., Lee, C. C. & Ribbe, M. W. Radical SAM-dependent carbon insertion into the nitrogenase M-cluster. Science 337, 1672–1675 (2012).
Wiig, J. A., Hu, Y. & Ribbe, M. W. Refining the pathway of carbide insertion into the nitrogenase M-cluster. Nat. Commun. 6, 8034 (2015).
Hu, Y. et al. FeMo cofactor maturation on NifEN. Proc. Natl Acad. Sci. USA 103, 17119–17124 (2006).
Hu, Y. et al. Nitrogenase Fe protein: a molybdate/homocitrate insertase. Proc. Natl Acad. Sci. USA 103, 17125–17230 (2006).
Ribbe, M. W., Hu, Y., Hodgson, K. O. & Hedman, B. Biosynthesis of nitrogenase metalloclusters. Chem. Rev. 114, 4063–4080 (2014).
Fay, A. W., Wiig, J. A., Lee, C. C. & Hu, Y. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc. Natl Acad. Sci. USA 112, 14829–14833 (2015).
Wilcoxen, J. et al. Electron paramagnetic resonance characterization of three iron-sulfur clusters present in the nitrogenase cofactor maturase NifB from Methanocaldococcus infernus. J. Am. Chem. Soc. 138, 7468–7471 (2016).
Averill, B. A., Herskovitz, T., Holm, R. H. & Ibers, J. A. Synthetic analogs of the active sites of iron–sulfur proteins. II. Synthesis and structure of the tetra(mercapto-μ3-sulfido-iron) clusters, (Fe4S4(SR)4)2–. J. Am. Chem. Soc. 95, 3523–3534 (1973).
Barclay, J. E., Davies, S. C., Evans, D. J. & Hughes, D. L. Lattice effects in the Mössbauer spectra of salts of [Fe4S4{S(CH2)nOH}4]2−. Crystal structures of [PPh4]2[Fe4S4{S(CH2)nOH}4] (n = 2, 3 and 4). Inorg. Chim. Acta 291, 101–108 (1999).
de Carvalho, L. M. & Schwedt, G. Sulfur speciation by capillary zone electrophoresis. Determination of dithionite and its decomposition products sulfite, sulfate and thiosulfate in commercial bleaching agents. J. Chromatogr. A 1099, 185–190 (2005).
Münchow, V. & Steudel, R. The decomposition of aqueous dithionite and its reactions with polythionates SnO2− 6 (n = 3–5) studied by ion-pair chromatography. Z. Anorg. Allg. Chem. 620, 121–126 (1994).
Vincent, K. A. et al. Instantaneous, stoichiometric generation of powerfully reducing states of protein active sites using Eu(ii) and polyaminocarboxylate ligands. Chem. Commun. 2003, 2590–2591 (2003).
Spatzal, T., Perez, K. A., Howard, J. B. & Rees, D. C. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron–molybdenum cofactor. Elife 4, e11620 (2015).
Spatzal, T., Perez, K. A., Einsle, O., Howard, J. B. & Rees, D. C. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science 345, 1620–1623 (2014).
Brychkova, G., Grishkevich, V., Fluhr, R. & Sagi, M. An essential role for tomato sulfite oxidase and enzymes of the sulfite network in maintaining leaf sulfite homeostasis. Plant Physiol. 161, 148–164 (2013).
Carbonero, F., Benefiel, A. C., Alizadeh-Ghamsari, A. H. & Gaskins, H. R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 3, 448 (2012).
Kertesz, M. A. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175 (2000).
Integrated Microbial Genomes (Joint Genome Institute); https://img.jgi.doe.gov/cgi-bin/m/main.cgi.
Mishanina, T. V., Libiad, M. & Banerjee, R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11, 457–464 (2015).
Frey, P. A., Hegeman, A. D. & Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008).
Mühlenhoff, U. et al. Compartmentalization of iron between mitochondria and the cytosol and its regulation. Eur. J. Cell Biol. 94, 292–308 (2015).
Mettert, E. L. & Kiley, P. J. Fe–S proteins that regulate gene expression. Biochim. Biophys. Acta 1853, 1284–1293 (2015).
O’Brien, E. et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355, eaag1789 (2017).
Berggren, G. et al. Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature 499, 66–69 (2013).
Esselborn, J. et al. Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat. Chem. Biol. 9, 607–609 (2013).
Shima, S. et al. Reconstitution of [Fe] hydrogenase using model complexes. Nat. Chem. 7, 995–1002 (2015).
Tanifuji, K. et al. Combining a nitrogenase scaffold and a synthetic compound into an artificial enzyme. Angew. Chem. Int. Ed. 54, 14022–14025 (2015).
Heinisch, T. & Ward, T. R. Artificial metalloenzymes based on the biotin–streptavidin technology: challenges and opportunities. Acc. Chem. Res. 49, 1711–1721 (2016).
Lin, Y.-W. Rational design of metalloenzymes: from single to multiple active sites. Coord. Chem. Rev. 336, 1–27 (2017).
Acknowledgements
This work was supported by NIH-NIGMS grant GM67626 (to M.W.R. and Y.H.), DOE-BES grant DE-DC0014470 (to M.W.R. and Y.H.), a Takeda Science Foundation grant (to Y.O.) and Grant-in-Aids for Scientific Research (nos 23000007 and 16H04116) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to K.Tat. and Y.O.).
Author information
Authors and Affiliations
Contributions
K.Tan., C.C.L., N.S.S., Y.H. and M.W.R. designed and analysed experiments. K.Tan. performed experiments. K.Tat. and Y.O. provided materials. Y.H. and M.W.R. wrote the manuscript with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, References, and Figures 1–6
Rights and permissions
About this article
Cite this article
Tanifuji, K., Lee, C.C., Sickerman, N.S. et al. Tracing the ‘ninth sulfur’ of the nitrogenase cofactor via a semi-synthetic approach. Nature Chem 10, 568–572 (2018). https://doi.org/10.1038/s41557-018-0029-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0029-4
This article is cited by
-
Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nature Catalysis (2022)
-
Tracing the incorporation of the “ninth sulfur” into the nitrogenase cofactor precursor with selenite and tellurite
Nature Chemistry (2021)
-
Bioassembly of complex iron–sulfur enzymes: hydrogenases and nitrogenases
Nature Reviews Chemistry (2020)
-
Identity and function of an essential nitrogen ligand of the nitrogenase cofactor biosynthesis protein NifB
Nature Communications (2020)
-
Probing the coordination and function of Fe4S4 modules in nitrogenase assembly protein NifB
Nature Communications (2018)