Abstract
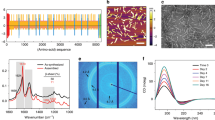
Post-translational modification of proteins is a strategy widely used in biological systems. It expands the diversity of the proteome and allows for tailoring of both the function and localization of proteins within cells as well as the material properties of structural proteins and matrices. Despite their ubiquity in biology, with a few exceptions, the potential of post-translational modifications in biomaterials synthesis has remained largely untapped. As a proof of concept to demonstrate the feasibility of creating a genetically encoded biohybrid material through post-translational modification, we report here the generation of a family of three stimulus-responsive hybrid materials—fatty-acid-modified elastin-like polypeptides—using a one-pot recombinant expression and post-translational lipidation methodology. These hybrid biomaterials contain an amphiphilic domain, composed of a β-sheet-forming peptide that is post-translationally functionalized with a C14 alkyl chain, fused to a thermally responsive elastin-like polypeptide. They exhibit temperature-triggered hierarchical self-assembly across multiple length scales with varied structure and material properties that can be controlled at the sequence level.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Langer, R. & Tirrell, D. A. Designing materials for biology and medicine. Nature 428, 487–492 (2004).
Maskarinec, S. A. & Tirrell, D. A. Protein engineering approaches to biomaterials design. Curr. Opin. Biotechnol. 16, 422–426 (2005).
Chilkoti, A., Dreher, M. R. & Meyer, D. E. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv. Drug Deliv. Rev. 54, 1093–1111 (2002).
Haider, M., Megeed, Z. & Ghandehari, H. Genetically engineered polymers: status and prospects for controlled release. J. Control. Release 95, 1–26 (2004).
Chow, D., Nunalee, M. L., Lim, D. W., Simnick, A. J. & Chilkoti, A. Peptide-based biopolymers in biomedicine and biotechnology. Mater. Sci. Eng. R. Rep. 62, 125–155 (2008).
Hochkoeppler, A. Expanding the landscape of recombinant protein production in Escherichia coli. Biotechnol. Lett. 35, 1971–1981 (2013).
Mann, M. & Jensen, O. N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Walsh, C. T., Garneau-Tsodikova, S. & Gatto, G. J. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. 44, 7342–7372 (2005).
Wold, F. In vivo chemical modification of proteins. Annu. Rev. Biochem. 50, 783–814 (1981).
Walsh, G. & Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252 (2006).
Pinkas, D. M., Ding, S., Raines, R. T. & Barron, A. E. Tunable, post-translational hydroxylation of collagen domains in Escherichia coli. ACS Chem. Biol. 6, 320–324 (2011).
Lim, S. et al. In vivo post-translational modifications of recombinant mussel adhesive protein in insect cells. Biotechnol. Prog. 27, 1390–1396 (2011).
Gordon, J. I., Duronio, R. J., Rudnick, D. A., Adams, S. P. & Gokel, G. W. Protein N-myristoylation. J. Biol. Chem. 266, 8647–8650 (1991).
Berndt, P., Fields, G. B. & Tirrell, M. Synthetic lipidation of peptides and amino acids: Monolayer structure and properties. J. Am. Chem. Soc. 117, 9515–9522 (1995).
Hartgerink, J. D., Beniash, E. & Stupp, S. I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 294, 1684–1688 (2001).
Hamley, I. W. Self-assembly of amphiphilic peptides. Soft Matter 7, 4122 (2011).
Cui, H., Webber, M. J. & Stupp, S. I. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers 94, 1–18 (2010).
Silva, G. A. et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science 303, 1352–1355 (2004).
Urry, D. W. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J. Phys. Chem. B 101, 11007–11028 (1997).
Roberts, S., Dzuricky, M. & Chilkoti, A. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 589, 2477–2486 (2015).
Urry, D. W. et al. Elastic protein-based polymers in soft tissue augmentation and generation. J. Biomater. Sci. Polym. Ed. 9, 1015–1048 (1998).
MacEwan, S. R. & Chilkoti, A. Elastin-like polypeptides: biomedical applications of tunable biopolymers. Biopolymers 94, 60–77 (2010).
Amiram, M., Luginbuhl, K. M., Li, X., Feinglos, M. N. & Chilkoti, A. Injectable protease-operated depots of glucagon-like peptide-1 provide extended and tunable glucose control. Proc. Natl Acad. Sci. USA 110, 2792–2797 (2013).
Luginbuhl, K. M. et al. One-week glucose control via zero-order release kinetics from an injectable depot of glucagon-like peptide-1 fused to a thermosensitive biopolymer. Nat. Biomed. Eng. 1, 0078 (2017).
McHale, M. K., Setton, L. A. & Chilkoti, A. Synthesis and in vitro evaluation of enzymatically cross-linked elastin-like polypeptide gels for cartilaginous tissue repair. Tissue Eng. 11, 1768–1779 (2005).
Lim, D. W., Nettles, D. L., Setton, L. A. & Chilkoti, A. In situ cross-linking of elastin-like polypeptide block copolymers for tissue repair. Biomacromolecules 9, 222–230 (2008).
MacEwan, S. R. & Chilkoti, A. Digital switching of local arginine density in a genetically encoded self-assembled polypeptide nanoparticle controls cellular uptake. Nano Lett. 12, 3322–3328 (2012).
Cho, Y. et al. Effects of Hofmeister anions on the phase transition temperature of elastin-like polypeptides. J. Phys. Chem. B 112, 13765–13771 (2008).
McDaniel, J. R., Radford, D. C. & Chilkoti, A. A unified model for de novo design of elastin-like polypeptides with tunable inverse transition temperatures. Biomacromolecules 14, 2866–2872 (2013).
Duronio, R. J. et al. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc. Natl Acad. Sci. USA 87, 1506–1510 (1990).
Heal, W. P. et al. Site-specific N-terminal labelling of proteins in vitro and in vivo using N-myristoyl transferase and bioorthogonal ligation chemistry. Chem. Commun. 3, 480–482 (2008).
Kulkarni, C., Kinzer-Ursem, T. L. & Tirrell, D. A. Selective functionalization of the protein N terminus with N-myristoyl transferase for bioconjugation in cell lysate. ChemBioChem 14, 1958–1962 (2013).
Kulkarni, C., Lo, M., Fraseur, J. G., Tirrell, D. A. & Kinzer-Ursem, T. L. Bioorthogonal chemoenzymatic functionalization of calmodulin for bioconjugation applications. Bioconjug. Chem. 26, 2153–2160 (2015).
Ho, S. H. & Tirrell, D. A. Chemoenzymatic labeling of proteins for imaging in bacterial cells. J. Am. Chem. Soc. 138, 15098–15101 (2016).
Eisenhaber, F. et al. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Π, NMT and PTS1. Nucleic Acids Res. 31, 3631–3634 (2003).
Paramonov, S. E., Jun, H. W. & Hartgerink, J. D. Self-assembly of peptide-amphiphile nanofibers: the roles of hydrogen bonding and amphiphilic packing. J. Am. Chem. Soc. 128, 7291–7298 (2006).
Ortony, J. H. et al. Internal dynamics of a supramolecular nanofibre. Nat. Mater. 13, 812–816 (2014).
Maurer-Stroh, S., Eisenhaber, B. & Eisenhaber, F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 317, 523–540 (2002).
Maurer-Stroh, S., Eisenhaber, B. & Eisenhaber, F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317, 541–557 (2002).
Römer, L. & Scheibel, T. The elaborate structure of spider silk: structure and function of a natural high performance fiber. Prion 2, 154–161 (2008).
Xu, X. D., Jin, Y., Liu, Y., Zhang, X. Z. & Zhuo, R. X. Self-assembly behavior of peptide amphiphiles (PAs) with different length of hydrophobic alkyl tails. Colloids Surf. B 81, 329–335 (2010).
Lee, O.-S., Stupp, S. I. & Schatz, G. C. Atomistic molecular dynamics simulations of peptide amphiphile self-assembly into cylindrical nanofibers. J. Am. Chem. Soc. 133, 3677–3683 (2011).
Smith, C. K., Withka, J. M. & Regan, L. A thermodynamic scale for the beta-sheet forming tendencies of the amino acids. Biochemistry 33, 5510–5517 (1994).
Minor, D. L. & Kim, P. S. Measurement of the [beta]-sheet-forming propensities of amino acids. Nature 367, 660–663 (1994).
Quiroz, F. G. & Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 14, 1164–1171 (2015).
Hassouneh, W., Christensen, T. & Chilkoti, A. Elastin-like polypeptides as a purification tag for recombinant proteins. Curr. Protoc. Protein Sci. 6, 6.11 (2010).
Meyer, D. E. & Chilkoti, A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules 3, 357–367 (2002).
Serrano, V., Liu, W. & Franzen, S. An infrared spectroscopic study of the conformational transition of elastin-like polypeptides. Biophys. J. 93, 2429–2435 (2007).
Jiang, H., Guler, M. O. & Stupp, S. I. The internal structure of self-assembled peptide amphiphiles nanofibers. Soft Matter 3, 454–462 (2007).
Cui, H., Cheetham, A. G., Pashuck, E. T. & Stupp, S. I. Amino acid sequence in constitutionally isomeric tetrapeptide amphiphiles dictates architecture of one-dimensional nanostructures. J. Am. Chem. Soc. 136, 12461–12468 (2014).
LeVine, H. 3rd Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 2, 404–410 (1993).
Stsiapura, V. I. et al. Thioflavin T as a molecular rotor: fluorescent properties of thioflavin T in solvents with different viscosity. J. Phys. Chem. B 112, 15893–15902 (2008).
Sulatskaya, A. I., Maskevich, A. A., Kuznetsova, I. M., Uversky, V. N. & Turoverov, K. K. Fluorescence quantum yield of thioflavin T in rigid isotropic solution and incorporated into the amyloid fibrils. PLoS ONE 5, e15385 (2010).
Newcomb, C. J., Moyer, T. J., Lee, S. S. & Stupp, S. I. Advances in cryogenic transmission electron microscopy for the characterization of dynamic self-assembling nanostructures. Curr. Opin. Colloid Interface Sci. 17, 350–359 (2012).
Mcdaniel, J. R. et al. Noncanonical self-assembly of highly asymmetric genetically encoded polypeptide amphiphiles into cylindrical micelles. Nano Lett. 14, 6590–6598 (2014).
Simon, J. R., Carroll, N. J., Rubinstein, M., Chilkoti, A. & Lopez, G. P. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat. Chem. 9, 509–515 (2017).
Muiznieks, L. D. & Keeley, F. W. Proline periodicity modulates the self-assembly properties of elastin-like polypeptides. J. Biol. Chem. 285, 39779–39789 (2010).
Muiznieks, L. D. et al. Modulated growth, stability and interactions of liquid-like coacervate assemblies of elastin. Matrix Biol. 36, 39–50 (2014).
Reichheld, S. E., Muiznieks, L. D., Keeley, F. W. & Sharpe, S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl Acad. Sci. USA 114, E4408–E4415 (2017).
Albertazzi, L. et al. Probing exchange pathways in one-dimensional aggregates with super-resolution microscopy. Science 344, 491–495 (2014).
da Silva, R. M. P. et al. Super-resolution microscopy reveals structural diversity in molecular exchange among peptide amphiphile nanofibres. Nat. Commun. 7, 11561 (2016).
Aluri, S., Pastuszka, M. K., Moses, A. S. & MacKay, J. A. Elastin-like peptide amphiphiles form nanofibers with tunable length. Biomacromolecules 13, 2645–2654 (2012).
Mejuch, T. & Waldmann, H. Synthesis of lipidated proteins. Bioconjug. Chem. 27, 1771–1783 (2016).
Triola, G., Waldmann, H. & Hedberg, C. Chemical biology of lipidated proteins. ACS Chem. Biol. 7, 87–99 (2012).
Inostroza-Brito, K. E. et al. Co-assembly, spatiotemporal control and morphogenesis of a hybrid protein–peptide system. Nat. Chem. 7, 897–904 (2015).
Acknowledgements
This research was funded by the National Science Foundation (NSF) through the Research Triangle Materials Research Science and Engineering Center (MRSEC; DMR-1121107) and by the National Institutes of Health (NIH; R01 GM-061232). Duke University Shared Materials Instrumentation Facility (SMIF) and Analytical Instrumentation facility (AIF) at North Carolina State University are members of the North Carolina Research Triangle NanotechnologyNetwork (RTNN), which is supported by the NSF (ECCS-1542015) as part of the National Nanotechnology Coordinated Infrastructure (NNCI). The authors thank K. Franz for access to chromatography instrumentation and the peptide synthesizer, J.G. Mark for conducting SDCLM experiments and M. Plue for the SEM imaging. The authors also thank H. Burg for her support in SFM sample preparation and analysis, M.-J. van Zadel for assistance with the variable-temperature ATR-IR experiments and M. Rubinstein for suggestions regarding the mechanism of self-assembly.
Author information
Authors and Affiliations
Contributions
D.M., K.M.L. and A.C. designed and performed experiments, analysed data and wrote the manuscript. J.R.S., M.D., R.B., H.S.V., S.H.P., F.C.H., K.L.B., N.R.M., I.W. and M.B. performed experiments, analysed data and took part in discussions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, data, analysis; Supplementary figures 1–4; Supplementary tables 1–6
Rights and permissions
About this article
Cite this article
Mozhdehi, D., Luginbuhl, K.M., Simon, J.R. et al. Genetically encoded lipid–polypeptide hybrid biomaterials that exhibit temperature-triggered hierarchical self-assembly. Nature Chem 10, 496–505 (2018). https://doi.org/10.1038/s41557-018-0005-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0005-z
This article is cited by
-
The construction of elastin-like polypeptides and their applications in drug delivery system and tissue repair
Journal of Nanobiotechnology (2023)
-
Applications of synthetic biology in medical and pharmaceutical fields
Signal Transduction and Targeted Therapy (2023)
-
Clickable, Oxidation-Responsive and Enzyme-Degradable Polypeptide: Synthesis, Characterization and Side Chain Modification
Chinese Journal of Polymer Science (2022)
-
Design of gel-to-sol UCST transition peptides by controlling polypeptide β-sheet nanostructures
Polymer Journal (2021)
-
Disordered protein-graphene oxide co-assembly and supramolecular biofabrication of functional fluidic devices
Nature Communications (2020)