Abstract
There is significant potential for employing algae in tertiary wastewater treatment, however, little is known about the contribution of algae-bacteria synergy toward treatment performance. This study demonstrates potential synergy in the treatment of three winery wastewater samples. Two strains of green algae, Auxenochlorella protothecoides and Chlorella sorokiniana were tested and each removed > 90% of nitrogen, > 50% of phosphate, and 100% of acetic acid in the wastewater. Both algae strains grew significantly faster on wastewaters compared to growth on minimal media. Organic carbon in the wastewater apparently played a limited role in algal growth enhancement. When cultured on sterile-filtered wastewater, A. protothecoides increased soluble COD loadings in two of the three wastewaters and C. sorokiniana secreted an insoluble film. Culturing algae with the native wastewater microbial community negated the secretion of algal photosynthate, allowing for simultaneous reductions in COD and nutrient concentrations. Both algae species stimulated bacterial growth in a strain-specific way, suggesting unique responses to algal photosynthate. Cofactor auxotrophy for thiamine, cobalamin, and biotin is widespread among algae and these cofactors are typically obtained from bacteria. Sequencing the wastewater microbial community revealed bacteria capable of synthesizing all three cofactors while liquid chromatography with mass spectrometry (LCMS) and bio-assays revealed the presence of thiamine metabolites in the wastewaters. These cofactors likely increased algal growth rates, particularly for A. protothecoides, which cannot synthesize thiamine de-novo but can salvage it from degradation products. Collectively, these results demonstrate that bacteria and algae provided synergistic growth benefits, potentially contributing to higher levels of wastewater treatment than either organism type alone.
Similar content being viewed by others
Introduction
Many agricultural and food processing operations are increasingly employing anaerobic digestion to reduce organic carbon loads in wastewaters while producing methane-rich biogas.1 However, digesters do a poor job of removing inorganic nutrients such as ammonium and phosphate, which are largely retained in the effluent, and can pollute waterways, contributing to harmful algal blooms.2 Compared to digesters, algae are not particularly effective at degrading complex organic molecules but are very effective at sequestering nitrogen and phosphorus from wastewaters, including those from anaerobic digesters.3,4 Algal biomass cultivated on wastewater can also be used for biofuel and bioproduct synthesis.5,6 Given the complimentary nature of these two technologies, cultivation of algae on anaerobic digester effluent could lead to high-quality wastewater effluent that has low concentrations of both organic and nutrient pollutants. High-quality effluent holds the potential for re-use in irrigation.
Numerous studies exist on the use of algae to treat various types of wastewaters including those from dairies,7 piggeries,8 wineries,9 slaughterhouses,10 and municipalities.11,12 The fact that algae can grow successfully on such chemically diverse wastewaters indicates the potential scalability of algal wastewater treatment. However, significant hurdles remain including potential negative interactions among native wastewater organisms and algae. The vast majority of studies on algal wastewater treatment focus on simple reporting of reductions in nutrient loads with little investigation into why a particular algae strain or mixed culture was effective. Moreover, numerous studies fail to explain if the wastewater was sterilized prior to algae cultivation; thus it is difficult to determine the relative contributions of algae and bacteria toward wastewater treatment performance.
In addition to measuring changes in water quality parameters in the present study, we also investigated how green algae and native wastewater organisms interact in the treatment process. Our past research has focused on interactions between green algae and bacteria in synthetic co-cultures. Thus, we were interested in understanding the extent to which our co-culture findings apply to a more complex wastewater environment. In previous studies, co-culturing the model organisms Escherichia coli and Auxenochlorella protothecoides led to synergistic interaction that increased algal growth rates and lipid production under mixotrophic conditions.13,14 Those same studies also showed that co-cultures could more rapidly remove organic and nitrogen species from the medium, potentially benefiting wastewater treatment. Further study revealed that much of the synergy was due to exchange of particular thiamine metabolites between the bacteria and algae.15 We hypothesized that similar synergy exists among many potential pairs of organisms due to widespread cofactor auxotrophy in algae16 and the important role of bacteria in providing these cofactors to algae in nature.17 In the present study, the potential for cofactor symbiosis between green algae and organisms present in winery wastewater was investigated. The relative contributions of algae and the native wastewater community toward the observed changes in water quality parameters were also studied.
Results
Algal wastewater treatment
Two strains of green algae, A. protothecoides and C. sorokiniana, were cultured in photobioreactors filled with sterile-filtered winery wastewater collected at three different times during the grape crush season. These wastewaters were categorized as “pre-crush,” “crush,” and “post-crush” in correspondence to the time in the wine production season when they were collected relative to the production of wine grape juice (grape crush). The winery employed a digester-based treatment system that yielded a wastewater low in organic content but high in nutrients, particularly nitrogen, phosphate, and sulfate (Table 1). The majority of nitrogen was in the form of ammonium. When cultured on sterile-filtered wastewater, both algae strains were effective at removing > 90% of total nitrogen, > 50% of phosphate, > 12% of sulfate, and all detectable acetic acid from the three wastewater types. C. sorokiniana in particular could remove all detectable nitrogen and phosphate in the crush and post-crush wastewaters within a span of 4 days. A. protothecoides could not metabolize the small quantities of nitrite and nitrate found in the pre-crush wastewater but was generally very effective at ammonium removal ( > 90%).
When A. protothecoides was grown on sterile-filtered wastewater, soluble organic material was secreted into the wastewater as observed by the increase in chemical oxygen demand (COD) loads in the pre- and post-crush wastewaters after algae cultivation. These compounds were also likely secreted into the crush wastewater but the net effect on COD was over-shadowed by algal consumption of acetate in the wastewater. C. sorokiniana also secreted material but it appeared to be polymeric in nature, forming a white film that pelleted with the algal cells and thus was not captured by the soluble COD assay.
The effect of algal treatment on removal of ions that contribute to salinity including chloride, sodium, potassium, magnesium, and calcium was modest or non-existent. Potassium removal was statistically significant for A. protothecoides cultures where removal levels were consistently between 30–50 mg/L regardless of the wastewater source. In the pre-crush wastewater, A. protothecoides appeared to increase the calcium concentration, however, this effect was more likely due to limited instrument sensitivity at such low concentrations.
Growth rates of algae on wastewater
All three wastewater types supported robust algae growth (Fig. 1a). In all cases, algae grew more rapidly on sterile-filtered wastewaters than they did on minimal media (controls). In the case of A. protothecoides, growth rates on wastewater were more than 13-fold greater than that on N8-NH4 minimal medium. N8-NH4 medium contains 257 mg/L ammonium, which is 1.3–2.2-fold higher than that found in the three wastewaters. C. sorokiniana growth on wastewater was roughly double that on N8 minimal medium, which contained 614 mg/L as nitrate. More detail on minimal media composition can be found in the Methods section.
Growth of algae and bacteria on wastewater. a Average dry weight growth rate of A. protothecoides over 5 days and C. sorokiniana over 4 days using sterile-filtered pre-crush, crush, and post-crush winery wastewaters. Control cultures were grown on minimal media under autotrophic conditions. Parts b–e show microbial (MC) and algal growth on post-crush wastewater. b Cultivation of A. protothecoides on minimal medium (control), on sterile-filtered wastewater, and on raw wastewater with the native microbial community. The far right bar shows net growth of the native microbial community without algae. c Cultivation of C. sorokiniana under the same treatments as described for A. protothecoides. d Total cultureable non-algae organisms based on plate counts in the presence and absence of A. protothecoides. e Cultureable organisms in the presence and absence of C. sorokiniana. For all panels, error bars denote standard deviation (n = 3 for post-crush and n = 4 for pre-crush and crush wastewater). Lowercase letters denote significance at the 0.05 level for algae, whereas uppercase letters are used for comparison of bacterial populations
Algal–bacterial complementarity in wastewater treatment
Thus far, results pertain to algae cultivated on sterile-filtered wastewater in order to understand algal interaction with chemical constituents in the wastewater. However, real treatment systems will include a complex mixture of bacteria along with algae. Additional culture studies were carried out to better understand the impact of the native wastewater microbial community on algal growth and wastewater treatment performance. In this experiment, algae were cultured on both sterile-filtered wastewater and on raw un-sterilized wastewater. Additional reactors were filled with raw wastewater without algae to understand the role of the native bacterial community in wastewater treatment performance. Acting alone, the bacterial community further reduced the COD loads in the wastewater by 36–60% over a 5-day period (Table 2). However, this community had significantly lower removal rates for nitrogen, phosphate, and sulfate compared to algae. By culturing the bacteria together with algae, significant reductions in both COD and nutrients were achieved. Significantly, the bacterial community appeared capable of consuming the organic photosynthate generated by A. protothecoides: culturing the algae on sterile-filtered wastewater significantly increased COD loads by 44% (p < 0.001), whereas the mixed culture reduced COD loads by 38% (p < 0.001). Neither the community nor algae were particularly effective at removing chloride, potassium, sodium, magnesium, or calcium ions.
Impact of algae on bacterial populations
Two approaches were utilized to quantify the total bacterial population in the post-crush wastewater: quantitative PCR and plate counts. The former quantifies total (living and dead) bacteria through quantitation of a segment of the 16S ribosomal RNA (rRNA) gene, and the latter captures information on viable counts of culturable organisms. Both methods showed that the presence of algae significantly increased the bacteria population size (Fig. 1b–e). Total bacterial growth rates based on PCR quantitation were ~ 44 mg/L/day in the presence of A. protothecoides and ~ 130 mg/L/day in the presence of C. sorokiniana compared to almost no detectable growth in the absence of algae (Fig. 1b,c). Likewise, Colony-forming units (CFU) counts of non-algae organisms were 4–7 times higher in treatments with algae than those without (Fig. 1d, e).
Cultivation of algae on post-crush wastewater resulted in faster growth than cultivation on chemical medium, regardless of whether filtered or raw wastewater was used. The presence of bacterial cells in the raw wastewater had no significant impact on A. protothecoides growth but led to a 23% decline in C. sorokiniana growth (Fig. 1b, c) compared to sterile-filtered wastewater. However, C. sorokiniana’s growth rate on raw wastewater was significantly higher than that on minimal medium.
To better understand the impact of aerobic treatment and algae on the microbial community in the raw wastewater, flow-cell sequencing of the V3–V4 region of the 16S rRNA gene was employed on the Illumina MiSeq platform. Mapping reads to operational taxonomic units (OTUs) revealed that the native wastewater community was composed primarily of organisms from the phyla Bacteroidetes, Firmicutes, Proteobacteria, and Synergistetes (Table 3). Cultivation of this community in the raw wastewater for 5 days in aerated bioreactors (without algae) led to a large shift in the microbial structure. This bioreactor-adapted community exhibited dramatic reduction in the relative abundances of Firmicutes and Synergistetes and an increase in Proteobacteria. When A. protothecoides was cultured in wastewater with the bacterial community, the relative abundances of Bacteroidetes and Actinobacteria were higher than the bioreactor-adapted community without algae. In co-culture with C. sorokiniana, the relative abundances of Bacteroidetes and Verrucomicrobia nominally increased but were not statistically significant. Given these changes, SIMPER analysis was performed using the Vegan package in R to determine OTUs that contributed most to dissimilarity among treatments. This revealed specific OTUs that increased in relative abundance in response to algae (Supplementary Table S1). In the presence of A. protothecoides, members of the genuses Sediminibacterium (OTU 7), Devosia (OTU 8), Luteolibacter (OUT 19), and of the order BD7-3 (OTU 16), all increased in relative abundance. In the presence of C. sorokiniana, a different set of organisms increased in relative abundance including members of Cryomorphaceae (OTU 3), Phenylobacterium (OTU 6), and BD7-3 (OTU 18).
We also searched the sequence data for signs of indicator bacterial pathogens, which is important for water re-use applications. The post-crush wastewater showed zero abundance for members of Escherichia, Salmonella, Shigella, and Vibrio. Very low relative abundance levels (<5) of Arcobacter were detected in the post-crush wastewater but were not detected after 5 days of culture in aerated bioreactors, regardless of the presence of algae.
Wastewater thiamine metabolites
We were interested in better understanding why algae grew more rapidly on wastewaters than they did on minimal medium. Past research has demonstrated that bacterial secretions can dramatically increase A. protothecoides growth through provision of thiamine metabolites. Given the diverse bacterial populations in the wastewaters and rapid A. protothecoides growth on wastewater, the presence of thiamine metabolites merited investigation. Two methods were used to determine if thiamine metabolites were present in the wastewaters: targeted liquid chromatography with mass spectrometry (LCMS) and a bio-assay. Direct measurement of five thiamine metabolites in the three wastewaters by targeted LCMS proved challenging due to instability of these metabolites in the wastewater. For instance, near complete destruction of thiamine monophosphate (TMP) and thiamine pyrophosphate (TPP) was observed within a few hours of exogenous addition of these metabolites to the wastewater (data not shown). Thiamine can degrade into a variety of products,18 of which, two potential candidates are 4-methyl-5-thiazoleethanol (THZ) and (4-amino-2-methyl-5-pyrimidine) methanol (HMP). Without accounting for the low recovery factors, quantifiable thiamine and HMP were observed in all three wastewaters. When correcting for recovery factors, concentrations of these two metabolites were in the range of 12–104 pM thiamine and 12–23 pM HMP (Table S2). These concentrations are far lower than the roughly 20 nM level required to fully alleviate thiamine limitation in A. protothecoides based on our previous experiments.15 With an observed A. protothecoides growth rate on wastewater of ~ 300 mg/L/day, some thiamine limitation is possible but previously published dose–response studies suggest thiamine metabolite concentrations on the order of 4 nM.15
In addition to LCMS analysis, a more general biological assay for total salvageable thiamine metabolites was carried out using A. protothecoides. Under thiamine-limited conditions, culture of A. protothecoides on glucose leads to secretion of pyruvate due to suppressed pyruvate dehydrogenase activity.15 A. protothecoides was, therefore, cultured in thiamine-free chemical medium and on all three sterilized wastewaters supplemented with 8 g/L glucose. Pyruvate secretion was measured by high-performance liquid chromatography (HPLC). The control culture secreted roughly 4 g/L pyruvate over the 5-day culture period. Wastewater cultures secreted minimal pyruvate (Fig. 2a). The crush wastewater resulted in a small (0.3 g/L) but statistically significant secretion of pyruvate (p = 0.045). These results suggest the presence of salvageable thiamine metabolites in all three wastewaters.
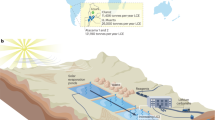
Thiamine metabolites in winery wastewater. a Bio-assay for useable thiamine metabolites. A. protothecoides was cultured on minimal medium and wastewaters supplemented with 8 g/L glucose. Glucose consumption and pyruvate secretion were measured after 5 days of culture. Error bars denote standard deviation (n = 3) and bars with the same letter are not significantly different at the 0.05 level. b Thiamine biosynthesis pathway using KEGG gene abbreviations. c Schematic of the wastewater treatment process employed at the winery. Wastewater from the plant’s crushing and washing operations is pumped to the equalization tank where the acidogenesis process begins. The anaerobic digesters utilize an expanded granular sludge bed to help retain biomass inside the digesters. Digester effluent then flows to an aerated oxidation tank prior to discharge to the local municipal wastewater treatment plant. This effluent was used for algae culture studies
Bacterial community cofactor synthesis capability
Based on a survey of 306 species, roughly half of all algae are likely auxotrophs for one or more B-vitamin cofactor, particularly cobalamin (B12), thiamine (B1), and biotin (B7).16 In nature, algae typically obtain these cofactors from exudates of bacteria, and B-vitamin abundance is governed by the size and composition of the bacterial community. To understand the capability of the wastewater microbial community to synthesize these three cofactors given their relevance to algae growth, the metagenomic prediction tool, PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), was applied to predict the likely abundance of key genes associated with synthesis pathways for thiamine (Fig. 2b), cobalamin (Supplementary Fig. S1), and biotin (Supplementary Fig. S2) within the wastewater microbial community.
PICRUSt analysis revealed the likely presence of complete biosynthesis pathways for all three B-vitamin cofactors within the microbial communities (Table 4, Supplementary Tables S3 and S4). Interestingly, the native microbial community typically exhibited the highest total abundance of cofactor synthesis genes, whereas the community adapted to the presence of algae typically had the lowest abundance. However, this trend was not consistent across all genes within each pathway.
An interesting result emerged in the analysis of cobalamin synthesis genes that revealed a shift from a partially anaerobic community to a more fully aerobic community after cultivation in the bioreactors. The native wastewater community had moderate abundance of genes corresponding to CbiE (K03399) and CbiT (K02191), which are unique to the anaerobic cobalamin synthesis pathway. Communities from the aerated bioreactors had almost no abundance of these genes but instead had high abundance of genes corresponding to CobN, CobS, and CobT, which are unique to the aerobic synthesis pathway. These results suggest a shift toward aerobic metabolism during culture in the bioreactors, regardless of the presence of algae.
Discussion
Culturing green algae with the native wastewater community resulted in the greatest removal of both COD and nutrient pollutants from the wastewater. In general, algae were effective at removing nutrients, whereas the microbial community consumed residual organic material including photosynthate secreted by algae. Ordinarily, algal photosynthate contributes to organic loads in wastewaters but the bacterial community could reverse this otherwise negative outcome, resulting in a net decrease in COD loads. This result is encouraging because it showed that not only were the algal and bacterial populations compatible but they also exhibited complementarity. However, none of the biological treatments could fully eliminate detectable soluble COD. The presence of algae also led to a significant increase in the total bacterial population size. This is further evidence that algal photosynthate likely provided organic carbon that supported bacterial growth. The shift in bacterial population composition in response to specific algae species also suggests an adaptation of the community to the specific photosynthate of each species. However, further research is required to characterize the photosynthate and elucidate bacterial metabolism of specific secretions.
One concern that may arise from this result is the potential for increased growth of wastewater pathogens due to the presence of algae. This was not specifically tested in this study. However, our previous result from co-culturing A. protothecoides with E. coli showed that E. coli populations were relatively unaffected by the presence of algae.13 Research on algal treatment of municipal wastewater has shown that algae cultivation can reduce coliform population size.19,20 It is also well-established that anaerobic digester treatment can suppress human pathogen population sizes,21,22 however, recent literature suggests that many pathogens enter a viable but non-culturable state during digestion.23 Thus, it would be important to implement a pathogen monitoring program for wastewater treated by anaerobic digestion followed by algae cultivation. This is particularly relevant if the water will be re-used in agricultural settings.
Acting alone or in conjunction with bacteria, algae were able to remove significant portions of nitrogen, phosphorus, and sulfur nutrients from the wastewaters. This finding is consistent with the results of other research efforts in which algal treatment led to complete or nearly complete nutrient removal from wastewater.8,9,10 Within this broad observation, an interesting trend emerged. The uptake ratios of total nitrogen and phosphate in algae cultures were nearly 3:1 in the pre-crush and crush wastewaters but were 1.8:1 in the post-crush wastewater. This was true for both algae species. The same N:P uptake ratio was observed in post-crush wastewater regardless of whether algae were cultured alone or in the presence of the microbial community. The difference in N:P uptake ratio is most likely due to the lower initial N:P ratio of the post-crush wastewater. The Redfield ratio suggests algal N:P ratios of 16:1 on a molar basis,24 which translates to a total nitrogen:phosphate ratio of 2.4. In reality, algae exhibit flexible nutrient uptake and elemental composition based on environmental conditions.25 For example, it is well-established that many algae species, including A. protothecoides and C. sorokiniana,14 respond to nitrogen limitation by reducing protein and increasing lipid production,26 thereby reducing the nitrogen content of their biomass.
In contrast to nutrient and organic matter removal, none of the biological treatments were effective at removing ions that frequently contribute to salinity such as sodium and chloride. Salinity is an important factor in cases where stakeholders, such as the collaborating winery in this study, wish to utilize wastewater for irrigation.27,28 The wastewaters in the present study were highly variable across different parts of the crush season with sodium concentrations varying from 97 to over 400 mg/L. Without substantial dilution, these concentrations generally exceed acceptable limits for fruit crop irrigation.29 The water may be acceptable for saline-tolerant crops such as sunflowers and sugar beets.29 Chloride concentrations were consistently <70 mg/L and are generally considered safe for all plants.
Given improved algae growth on wastewater compared to minimal medium, we were interested in understanding the source of this growth promotion. Faster-growing algae not only increase nutrient removal rates but also facilitate more cost-effective production of biofuels from wastewater-grown algae.30 Because growth enhancement could be achieved in sterile-filtered wastewater, (bio)chemical constituents in the wastewater were the likely source of growth promotion. Increased algae growth on wastewater is frequently attributed to mixotrophic growth due to organic material found in wastewaters.31 However, relatively low (<300 mg/L) COD levels were measured in the winery wastewaters. The crush wastewater had the highest COD level and also contained 344 mg/L residual acetic acid. Both algae strains consumed all acetate within 24 h but given the low concentrations, mixotrophy is unlikely to fully explain the robust algae growth on the three wastewaters, particularly in the case of A. protothecoides. In a previous study, the addition of 500 mg/L of acetate to an axenic A. protothecoides culture only increased growth rates by 2.4-fold13 compared to the roughly 13-fold increase observed on winery wastewater. Nutrient abundance also cannot explain the growth promotion, as the N8-NH4 minimal medium had higher ammonium and phosphate concentrations than all three wastewaters.
Our previous research on algal–bacterial interaction revealed important synergies related to thiamine metabolism that resulted in faster algae growth and lipid production.15 While the presence of live bacteria in wastewater did not improve algae growth in this study (and slightly repressed it in the case of C. sorokiniana), it is likely that bacterial exudates in the wastewater had a positive effect on algae growth, particularly in the case of A. protothecoides. Such exudates persist in water even after removal or death of the organisms that created them, thus co-culture with live bacterial cells is not necessarily a prerequisite for improved algae growth. Past research on a model co-culture system showed that exudates from E. coli contained thiamine metabolites, which could increase A. protothecoides growth rates 10–30-fold.15 That work also showed that A. protothecoides could obtain similar benefits from degraded thiamine products. Therefore, detecting thiamine metabolites in exudates from wastewater bacteria were of particular interest in the present study. The bio-assay and LCMS results suggest that the winery wastewaters contained utilizable thiamine metabolites, despite significant degradation. This further underscores our previous finding that A. protothecoides can salvage thiamine from degradation products. C. sorokiniana is not a thiamine auxotroph but did show modest growth enhancement when provided with exogenous thiamine in previous research.15 Thus provision of cofactors by the wastewater community could have partially contributed to the increased growth of this species on wastewater. Nevertheless, other mechanisms could also play a role in algal growth promotion on wastewater: bacterial provision of algal-growth-promoting hormones32,33 or the provision of other non-thiamine cofactors.
We did not specifically assay for other vitamin cofactors but predicted metagenomes from PICRUSt analysis suggest that the wastewater microbial communities contained complete synthesis pathways for thiamine, biotin, and cobalamin. These cofactors have been identified as particularly relevant to algae: in a survey of 306 algal species, 51% required cobalamin, 22% required thiamine, and 6% required biotin.16 In nature, algae typically obtain these cofactors from bacteria, as indicated by a number of studies.17,34 Paerl et al.35 demonstrated the potential of Pseudomonas sp. to transform TPP into a form useable by the green algae Ostreococcus lucimarinus, a known thiamine auxotroph. In another study, marine bacteria were shown to produce a thiazole precursor used by Ostreococcus species to synthesize thiamine.36 These findings are analogous to our past findings in co-cultures of E. coli with A. protothecoides and indicate the widespread importance of cofactor symbiosis in promoting algae growth.
Complete cofactor synthesis pathways in the microbial community may not be required to support auxotrophic algae growth due to the prevalence of partial synthesis and salvage pathways in algae such as those for thiamine mentioned previously. With regard to cobalamin, bacteria in consortia are known to remodel corrinoids obtained from other species,37 however, studies on algae are limited. One recent study has shown that some eukaryotic algae can convert pseudocobalamin into cobalamin when provided with 5,6-dimethylbenzimidazole.38 In light of salvage mechanisms, we predict that the winery wastewater could support a wide range of algae species, including a variety of B-vitamin auxotrophs, which are estimated to comprise half of algae species.16 This opens the possibility of engineering a process that employs specific algae species with desired co-product characteristics. It also allows for cultivation of algal polycultures, which hold the promise of greater process stability through species complementarity.39 Synergies between algae and bacteria should be a top consideration when designing suspended-cell processes for wastewater treatment.
Methods
Wastewater collection and preparation
Wastewater was obtained from a treatment plant located at a large winery in California (Fig. 2c). This on-site plant primarily treats wastewater from the winery’s crushing and tank washing operations. The effluent from the on-site treatment plant is then sent to a local municipal wastewater treatment plant for additional treatment at a significant cost to the winery.
Wastewaters were collected in clean plastic containers from a sampling port exiting the oxidation tank at the winery’s facility. Wastewater was kept on ice for transport back to the lab at UC Davis. Thereafter, wastewaters were immediately frozen at −20 °C until use. Thawed wastewater was used directly for the non-axenic cultures. Sterile wastewaters were prepared by centrifuge to remove large particulates followed by filtration first through a 0.45 μm filter and then through a 0.2 μm sterile filter to remove bacteria.
Strain selection
A. protothecoides (UTEX 2341)40 and C. sorokiniana (UTEX 2805) are model green algae and were selected for this study. Both strains grow rapidly and remove ammonium and phosphate from culture medium, important criteria for nutrient removal from wastewater. C. sorokiniana was isolated from a wastewater treatment pond in Mexico and is also capable of utilizing nitrate.41 C. sorokiniana accumulates intracellular starch, which has value for biofuel production from wastewater-grown algae.42 A. protothecoides accumulates neutral lipids and past work revealed that E. coli stimulated increased acetate and nutrient uptake by this strain.13,14 There was interest in whether wastewater bacteria could stimulate a similar response.
Culture experiments on wastewater
A series of culture experiments were carried out in which algae were cultured on sterile-filtered wastewater and also raw wastewater using the specific procedures described in the following section. In the first set of experiments, A. protothecoides and subsequently C. sorokiniana were cultured on sterile-filtered pre-crush, crush, and post-crush wastewater alongside autoclaved minimal medium as a control. These experiments revealed the “algae-only” contribution to wastewater treatment. Subsequently, the post-crush wastewater was selected for the study of algae growth on raw wastewater to better understand interactions with live bacteria. The reason post-crush wastewater was selected was to ensure that freshly collected wastewater was used, given the time-sensitive nature of its microbial community. In this round of culture experiments, first A. protothecoides and then C. sorokiniana were cultured on sterile filtered and raw post-crush wastewater. An additional set of reactors containing only raw wastewater (and its associated community) was also included in each of these experiments. Finally, algae cultured on autoclaved minimal medium served as a control for algae growth. Four biological replicate cultures were used per experimental treatment for pre-crush wastewaters. Three biological replicates per experimental treatment were used for post-crush wastewaters. These values were found to give sufficient power based on previous culture studies on winery wastewater. For all culture experiments, cultures were handled using sterile technique and samples were taken daily for optical density, pH, and HPLC analysis of the filtered (0.2 μm) wastewater. Additional samples were taken at culture harvest for total nitrogen and COD analysis. For cultures in raw wastewater, additional samples were taken on the first and last day of the culture period for viable bacteria enumeration by plating.
Algae and bacterial culture procedures
Algae pre-cultures were established in sterile 1 L bioreactors from colonies scraped from ATCC #5 agar plates. A. protothecoides was cultured on N8-NH4 medium13 and C. sorokiniana was cultured on N8 medium42 until cell density reached roughly OD550 of 0.2, which corresponds to 0.11 and 0.13 g/L for A. protothecoides and C. sorokiniana, respectively. Pre-cultures were supplied with 400 mL/min sterile-filtered 2% CO2 mixed with air and illuminated by T5 growth lamps (10,000 lux operating on a 16:8 light/dark cycle). Algae were allowed to settle in the bioreactor for 24 h and residual medium was removed first by aspiration and then by centrifugation and decanting of supernatant. Algae pre-cultures were then used to inoculate a set of 12 sterile 300 mL hybridization tubes containing 200 mL of various wastewaters or control media. Inoculation density was roughly OD550 of 0.16. Full factorial designs were used with a minimum of three biological replicates for each treatment.
Hybridization tubes were suspended in a temperature-controlled water bath (28 °C) and mixed by magnetic stir bar (~ 150 rpm). The same illumination scheme as the stock cultures was used. A mixture of 2% CO2 and air was sterile filtered and supplied to each reactor at a rate of 125 mL/min. Cultures were grown to log phase (4–5 days) and reactors were rotated to new incubator positions each day. At the end of the culture period, cells were harvested by centrifugation at 6000 rcf for 5 min. Algae harvested by centrifuge were washed three times with dH2O to remove medium components and then freeze-dried to obtain a final “harvestable” dry weight.
Viable bacterial counts in wastewater
For non-axenic algae cultured on raw post-crush wastewater, additional samples were taken on the first and last day of the culture period for plating on ATCC #5 agar. This agar contains a rich variety of metabolites from yeast extract, beef extract, and tryptose and can support a range of bacteria and algae. Samples were plated at a range of dilutions and CFUs were counted to determine viable and culturable organisms. Only non-algae species were tabulated (algae typically take > 5 days to appear on plates).
High-pressure liquid chromatography analysis of wastewater
HPLC and ion chromatography for wastewater analyses were carried out on a Shimadzu Prominence instrument (20 series) equipped with a 10 series pump. Organic acid analysis, including acetic and pyruvic acids, was carried out using an Aminex 87H column (Bio-Rad). Mobile phase was 5 mM sulfuric acid in MilliQ water with flow rate set to 0.6 mL/min. Shimadzu RID (10A) and PDA (20A) detectors were used. Suppressed cation chromatography was performed using a 4 × 250 mm CS14 column (Dionex), CERS500 suppressor, and Shimadzu conductivity detector (CDD-10AVP) based on the method described by Yu et al.43 Mobile phase was 10 mM methanesulfonic acid in MilliQ water with a flow rate of 1 mL/min. Anion chromatography was performed using a 4 × 250 mm AS22 column (Dionex), AERS500 suppressor, and conductivity detector. Mobile phase was a solution of 4.5 mM sodium carbonate and 1.5 mM sodium bicarbonate in MilliQ water. Chromatograms were analyzed in LC Solutions software (v. 1.23).
COD and nitrogen assays
Samples were collected at the beginning and end of the culture period and filtered (0.2 μm) to remove cells. Hach assay kits for COD (high range) and total nitrogen (low range) were used and sample vials were read in a DR/890 colorimeter. Procedures followed the manufacturer’s instructions; distilled water was used for blanks and dilutions.
Extraction of thiamine metabolites and targeted LCMS
Sterile-filtered wastewater (35 mL) was freeze-dried and extracted with methanol to recover thiamine metabolites per a previously established procedure.15 Recovery standards were prepared with known concentrations of five thiamine metabolites: thiamine-HCl, TMP, TPP, HMP, and THZ. Recovery rates for TMP, TPP, and THZ were low (Supplementary Table S1) so C18 solid-phase extraction approach was employed based on the method developed by Okbamichael and Sañudo-Wilhelmy for B-vitamin concentration in seawater.44 This method worked reasonably well for standards prepared in phosphate buffer but substantial losses of all metabolites were observed for recovery standards prepared in wastewater. Thus, the original methanol extraction method was used for the final analysis of thiamine and HMP, which had higher recovery efficiencies than the other metabolites.
Targeted LCMS analysis of thiamine metabolites in wastewater extracts was performed on a 1200 series LC (Agilent) coupled to an AB Sciex 4000 QTrap LC/MS/MS system. A HILIC method was used for separation and MRM setpoints were established to quantify thiamine, TMP, TPP, HMP, and THZ in the samples as described previously.15
Bio-assay for determination of useable thiamine metabolites
A bio-assay for useable thiamine was carried out using A. protothecoides. This organism secretes large quantities of pyruvate into the culture medium when cultured on glucose in the absence of useable thiamine metabolites. This is due to limited functionality of its pyruvate dehydrogenase complex in the absence of the cofactor TPP. For this study, A. protothecoides was grown on all three wastewaters, supplemented with 8 g/L glucose. As a positive control, it was also cultured on N8-NH4 medium (which contains no thiamine) supplemented with 8 g/L glucose. Culture samples were obtained daily, centrifuged, and the supernatant was filtered through 0.2 μm filters. Filtered samples were analyzed by HPLC to quantify glucose uptake and pyruvate secretion.
DNA extraction and qPCR for bacterial analysis
DNA was extracted from harvested cultures grown on post-crush wastewater. These cultures included raw wastewater prior to bioreactor culture, raw wastewater after bioreactor culture (without algae), and algae cultured in raw wastewater. DNA extraction was performed on harvested freeze-dried biomass using the Fast DNA Spin Kit (MP Biomedicals). Procedures followed recommendations in the kit. Specifically, 1.5 mg of freeze-dried biomass was suspended in 200 μL of distilled water and vortexed. Since the goal of this extraction was to recover bacterial DNA for quanitification and sequencing, 1 mL of the bacterial cell lysis buffer in the Fast DNA Spin Kit was then added to the biomass suspension. Lysing matrix A tubes were used to homogenize the samples twice in a FastPrep instrument (Savant) at 6 m/s for 30 s. Homogenized samples were incubated at room temperature for 30 min per the manufacturer’s recommendation for dry biomass. Remaining steps followed kit instructions.
A small portion (1 μL) of the extracted DNA was used to perform quantitative PCR on a segment of the 16S rRNA gene. The goal of this analysis was to quantify the total bacterial fraction of biomass in samples where algae were cultured in raw wastewater. Similar analysis on co-cultures of A. protothecoides and E. coli has been employed in past work with validated results.13 In that case, an E. coli-specific primer was used. In the present analysis, the goal was amplification of total bacterial 16S rRNA genes and thus only conserved gene regions could be used for primer development. The challenge with this approach is that amplification of conserved regions can also result in amplification of algal chloroplast genes. This contamination could be minimized through use of the 799F-mod3 (5′-CMGGATTAGATACCCKGG-3′) and 926 R (5′-CCGTCAATTCMTTTRAGTTT-3′) primers as recommended by Hanshew et al.45 The 799F-mod3 primer has been shown to effectively resist amplification of chloroplast 16S rRNA genes with < 1% chloroplast amplification in a 50–50 mock mixture of chloroplast and bacterial DNA.45 Quantitative PCR was carried out on a StepOnePlus instrument (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). Template DNA was diluted tenfold and 2 μL was added to the reaction tube. Reaction volume was 25 μL and primer concentrations were 100 nM. The thermocycler ran for 35 cycles: 95 °C for 15 s, 47 °C for 15 s, and 72 °C for 45 s. PCR product size (~ 130 bp) was confirmed by gel electrophoresis using a 2% agarose gel with a 100 bp ladder. Threshold cycle methodology was employed to quantify 16S rRNA genes using serial dilutions of previously amplified 16S rRNA gene product. In all, 16S rRNA gene quantity was correlated to the dry weight of bacterial cell mass in the raw wastewater (without algae). Using this correlation, the total bacterial population in the mixed alga-bacteria samples was quantified. One assumption of this approach is that the bacterial community was relatively unaffected by the presence of algae. Microbial community analysis revealed some statistically significant population shifts in response to algae but the magnitudes of the shifts were not dramatic (see Table 3).
16S rRNA gene sequencing and microbial community analysis
The remaining extracted DNA was submitted to CD Genomics (Shirley, NY) for flow-cell sequencing on an Illumina MiSEQ instrument. In all, 300 bp paired-end reads were obtained for the V3–V4 region (~ 250 bp) of the 16S rRNA gene. Reads were quality-filtered and demultiplexed by CD Genomics using a proprietary software pipeline. The resulting FASTA files were then loaded into QIIME (Quantitative Insights Into Microbial Ecology, v. 1.9.1) for removal of chimeric sequences with usearch v. 6.1. The cleaned FASTA file was then used for OTU picking against the Greengenes database (v. 13.5) at 97% identity. Two different OTU picking procedures were used: closed-reference OTU picking was used for subsequent PICRUSt analysis, whereas open-reference OTU picking was used for microbial community analysis. Supplementary Table S5 shows mapping rates for both methods. Singletons and chloroplast hits were removed from the dataset prior to further analysis. The Vegan package (v. 2.4–1)46 in R was used to calculate Shannon diversity, richness, and evenness of microbial communities. Vegan was also used to calculate the contributions of specific OTUs to Bray-Curtis dissimilarity using SIMPER analysis.
PICRUSt prediction of metagenomes
The metagenome prediction tool PICRUSt (v. 1.0.0) was used to infer gene functions available to the wastewater microbial community. This tool determines genes likely to be present in a reference OTU tree (from Greengenes) through ancestral state reconstruction and the use of a reference genome database.47 Closed-reference OTUs picked in QIIME were subsequently imported into PICRUSt. Because only 70% of the 209,580 total sequences mapped to reference OTUs at 97% identity, the PICRUSt analysis is likely to be conservative. One of the most important metrics for accurate metagenome predictions is the similarity between environmental organisms and the suite of reference genomes used in the PICRUSt algorithm. This dissimilarity can be quantified by the nearest sequence taxon index (NSTI) value with values below 0.18 resulting in reasonably accurate predictions.47 Of the 12 samples analyzed in this study, all but one had an NSTI below 0.18 with an average score of 0.15 (Supplementary Table S6).
Statistical analysis
Basic statistics including mean, standard deviation, and independent group 2-sided t-tests were performed in Microsoft Excel. ANOVA and multiple comparison tests were performed in R (v. 3.1.1)48 using the ‘‘car’’ and ‘‘agricolae’’ packages. Homogeneity of variance was tested using both Levene’s test and the Brown and Forsythe test. Where these tests showed significance, data were transformed using established methods49 to reduce heteroscedasticity prior to application of ANOVA. No data points from culture replicates were discarded from the datasets. Analytical assays were repeated additional times if an anomalous value was observed.
Data availability
This Targeted Locus Study project has been deposited at DDBJ/ENA/GenBank under the accessions KBPZ00000000, KBQA00000000, KBQB00000000, KBQC00000000, KBQD00000000, KBQE00000000, KBQF00000000, KBQG00000000, KBQH00000000, KBQI00000000, KBQJ00000000, KBQK00000000, KBQL00000000, KBQM00000000, KBQN00000000, KBQO00000000. The versions described in this paper are the first versions, KBPZ01000000, KBQA01000000, KBQB01000000, KBQC01000000, KBQD01000000, KBQE01000000, KBQF01000000, KBQG01000000, KBQH01000000, KBQI01000000, KBQJ01000000, KBQK01000000, KBQL01000000, KBQM01000000, KBQN01000000, KBQO01000000. In addition, the chimera-cleaned multi-line FASTA file obtained from 16S rRNA sequencing is publicly accessible at (https://www.dropbox.com/sh/a4ffenszaxx4ein/AAB7XIMJO0sHL9CAHJz8btSha?dl=0). Other data types are available upon request.
References
Goldstein, N. Agricultural anaerobic digestion in the US. Biocycle 54, 41 (2013).
Wilkie, A. C. & Mulbry, W. W. Recovery of dairy manure nutrients by benthic freshwater algae. Bioresour. Technol. 84, 81–91 (2002).
Woertz, I., Feffer, A., Lundquist, T. & Nelson, Y. Algae grown on dairy and municipal wastewater for simultaneous nutrient removal and lipid production for biofuel feedstock. J. Environ. Eng. 135, 1115–1122 (2009).
Franchino, M., Tigini, V., Varese, G. C., Mussat Sartor, R. & Bona, F. Microalgae treatment removes nutrients and reduces ecotoxicity of diluted piggery digestate. Sci. Total Environ. 569–570, 40–45 (2016).
Cole, A. J. et al. Adding value to the treatment of municipal wastewater through the intensive production of freshwater macroalgae. Algal Res. 20, 100–109 (2016).
Hodgskiss, L. H., Nagy, J., Barnhart, E. P., Cunningham, A. B. & Fields, M. W. Cultivation of a native alga for biomass and biofuel accumulation in coal bed methane production water. Algal Res. 19, 63–68 (2016).
Passero, M., Cragin, B., Coats, E. R., McDonald, A. G. & Feris, K. Dairy wastewaters for algae cultivation, polyhydroxyalkanote reactor effluent versus anaerobic digester effluent. BioEnergy Res. 8, 1647–1660 (2015).
Prandini, J. M. et al. Enhancement of nutrient removal from swine wastewater digestate coupled to biogas purification by microalgae Scenedesmus spp. Bioresour. Technol. 202, 67–75 (2016).
Liu, C. et al. Phycoremediation of dairy and winery wastewater using Diplosphaera sp. MM1. J. Appl. Phycol. 28, 3331–3341 (2016).
Hernández, D. et al. Microalgae cultivation in high rate algal ponds using slaughterhouse wastewater for biofuel applications. Chem. Eng. J. 285, 449–458 (2016).
Krustok, I., Odlare, M., Truu, J. & Nehrenheim, E. Inhibition of nitrification in municipal wastewater-treating photobioreactors: Effect on algal growth and nutrient uptake. Bioresour. Technol. 202, 238–243 (2016).
Bohutskyi, P. et al. Effects of inoculum size, light intensity, and dose of anaerobic digestion centrate on growth and productivity of Chlorella and Scenedesmus microalgae and their poly-culture in primary and secondary wastewater. Algal Res. 19, 278–290 (2016).
Higgins, B. & VanderGheynst, J. Effects of Escherichia coli on mixotrophic growth of Chlorella minutissima and production of biofuel precursors. PLoS ONE 9, e96807 (2014).
Higgins, B., Labavitch, J. M. & VanderGheynst, J. S. Co-culturing Chlorella minutissima with Escherichia coli can increase neutral lipid production and improve biodiesel quality. Biotechnol. Bioeng. 112, 1801–1809 (2015).
Higgins, B. T. et al. Cofactor symbiosis for enhanced algal growth, biofuel production, and wastewater treatment. Algal Res. 17, 308–315 (2016).
Croft, M. T., Warren, M. J. & Smith, A. G. Algae need their vitamins. Eukaryot. Cell 5, 1175–1183 (2006).
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J. & Smith, A. G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005).
Jenkins, A. H., Schyns, G., Potot, S., Sun, G. & Begley, T. P. A new thiamin salvage pathway. Nat. Chem. Biol. 3, 492–497 (2007).
Ansa, E. D. O., Lubberding, H. J. & Gijzen, H. J. The effect of algal biomass on the removal of faecal coliform from domestic wastewater. Appl. Water Sci. 2, 87–94 (2012).
Muñoz, R. & Guieysse, B. Algal–bacterial processes for the treatment of hazardous contaminants: A review. Water Res. 40, 2799–2815 (2006).
Dixo, N. G. H., Gambrill, M. P., Catunda, P. F. C. & van Haandel, A. C. Removal of pathogenic organisms from the effluent of an upflow anaerobic digester using waste stabilization ponds. Water Sci. Technol. 31, 275–284 (1995).
Kearney, T. E., Larkin, M. J., Frost, J. P. & Levett, P. N. Survival of pathogenic bacteria during mesophilic anaerobic digestion of animal waste. J. Appl. Bacteriol. 75, 215–219 (1993).
Fu, B., Jiang, Q., Liu, H. B. & Liu, H. Quantification of viable but nonculturable Salmonella spp. and Shigella spp. during sludge anaerobic digestion and their reactivation during cake storage. J. Appl. Microbiol. 119, 1138–1147 (2015).
Gruber, N. & Deutsch, C. A. Redfield’s evolving legacy. Nat. Geosci. 7, 853–855 (2014).
Planavsky, N. J. The elements of marine life. Nat. Geosci. 7, 855–856 (2014).
Piorreck, M., Baasch, K. -H. & Pohl, P. Biomass production, total protein, chlorophylls, lipids and fatty acids of freshwater green and blue-green algae under different nitrogen regimes. Phytochemistry 23, 207–216 (1984).
Muyen, Z., Moore, G. A. & Wrigley, R. J. Soil salinity and sodicity effects of wastewater irrigation in South East Australia. Agric. Water Manag. 99, 33–41 (2011).
Pedrero, F. & Alarcón, J. J. Effects of treated wastewater irrigation on lemon trees. Desalination 246, 631–639 (2009).
Bauder, T. A., Waskom, R. M., Sutherland, P. L. & Davis, J. G. Irrigation Water Quality Criteria. (Colorado State University, Fort Collins, CO, 2014).
Davis, R., Aden, A. & Pienkos, P. T. Techno-economic analysis of autotrophic microalgae for fuel production. Appl. Energy 88, 3524–3531 (2011).
Bhatnagar, A., Chinnasamy, S., Singh, M. & Das, K. C. Renewable biomass production by mixotrophic algae in the presence of various carbon sources and wastewaters. Appl. Energy 88, 3425–3431 (2011).
de-Bashan, L. E. Involvement of the indole-3-acetic acid produced by the growth-promoting bacterium Azospirillum spp. in promoting growth in Chlorella vulgaris. J. Phycol. 44, 938–947 (2008).
Bajguz, A. & Piotrowska-Niczyporuk, A. Synergistic effect of auxins and brassinosteroids on the growth and regulation of metabolite content in the green alga Chlorella vulgaris (Trebouxiophyceae). Plant Physiol. Biochem.: PPB / Soc. Fr. De. Physiol. Veg. 71, 290–297 (2013).
Kazamia, E. et al. Mutualistic interactions between vitamin B12 -dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 14, 1466–1476 (2012).
Paerl, R. W., Bertrand, E. M., Allen, A. E., Palenik, B. & Azam, F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol. Oceanogr. 60, 215–228 (2015).
Paerl, R. W. et al. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 11, 753–765 (2017).
Men, Y. et al. Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Appl. Environ. Microbiol. 80, 2133–2141 (2014).
Helliwell, K. E. et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B(12). Curr. Biol. 26, 999–1008 (2016).
Newby, D. T. et al. Assessing the potential of polyculture to accelerate algal biofuel production. Algal Res. 19, 264–277 (2016).
Higgins, B. T. et al. Informatics for improved algal taxonomic classification and research: A case study of UTEX 2341. Algal Res. 12, 545–549 (2015).
de-Bashan, L. E., Trejo, A., Huss, V. A. R., Hernandez, J. -P. & Bashan, Y. Chlorella sorokiniana UTEX 2805, a heat and intense, sunlight-tolerant microalga with potential for removing ammonium from wastewater. Bioresour. Technol. 99, 4980–4989 (2008).
Tanadul, O. U., Vandergheynst, J. S., Beckles, D. M., Powell, A. L. & Labavitch, J. M. The impact of elevated CO2 concentration on the quality of algal starch as a potential biofuel feedstock. Biotechnol. Bioeng. 111, 1323–1331 (2014).
Yu, C. et al. Influence of leaching pretreatment on fuel properties of biomass. Fuel Process. Technol. 128, 43–53 (2014).
Okbamichael, M. & Sañudo-Wilhelmy, S. A. Direct determination of vitamin B1 in seawater by solid-phase extraction and high-performance liquid chromatography quantification. Limnol. Oceanogr.: Methods 3, 241–246 (2005).
Hanshew, A. S., Mason, C. J., Raffa, K. F. & Currie, C. R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 95, 149–155 (2013).
Oksanen, J. et al. vegan: Community Ecology Package, <http://cran.r-project.org/package=vegan>(2015).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2017). https://www.R-project.org/.
Box, G. E. P. & Cox, D. R. An Analysis of Transformations. Journal of the Royal Statistical Society. Series B 26, 211–252 (1964).
Acknowledgements
This research was supported by NSF grants CBET-1438211 and MCB-1139644 and National Institute of Food and Agriculture project CA-D-BAE-2228-RR.
Author information
Authors and Affiliations
Contributions
B.T.H and J.S.V. designed the experiments. B.T.H. and S.J.C. conducted experiments. B.T.H. analyzed sequencing and LC datasets. I.G., P.S.F., and O.F. carried out LCMS analysis. B.T.H. wrote the manuscript. J.S.V. and S.J.C. also contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Higgins, B.T., Gennity, I., Fitzgerald, P.S. et al. Algal–bacterial synergy in treatment of winery wastewater. npj Clean Water 1, 6 (2018). https://doi.org/10.1038/s41545-018-0005-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41545-018-0005-y
This article is cited by
-
Euglena mutabilis exists in a FAB consortium with microbes that enhance cadmium tolerance
International Microbiology (2024)
-
Winery Wastewater Treatment: a Systematic Review of Traditional and Emerging Technologies and Their Efficiencies
Environmental Processes (2023)
-
Combined inorganic nitrogen sources influence the release of extracellular compounds that drive mutualistic interactions in microalgal‒bacterial co-cultures
Journal of Applied Phycology (2022)
-
Extracellular Polymeric Substances Drive Symbiotic Interactions in Bacterial‒Microalgal Consortia
Microbial Ecology (2022)
-
Impact of Nitrate and Ammonium Concentrations on Co-Culturing of Tetradesmus obliquus IS2 with Variovorax paradoxus IS1 as Revealed by Phenotypic Responses
Microbial Ecology (2022)