Abstract
Serum bilirubin is associated with several clinical outcomes, including hypertension, type 2 diabetes (T2D), and drug metabolism. Here, we describe findings from our genome-wide association studies (GWAS) of serum (TBIL) using a generalized linear mixed model in West Africans (n = 1127), with adjustment for age, sex, body mass index, T2D, significant principal components of population structure, and cryptic relatedness. Genome-wide conditional analysis and CAVIARBF were used to fine map significant loci. The causal effect of TBIL on hypertension was assessed by Mendelian randomization (MR) using the GWAS findings as instrumental variables (IVs) in African Americans (n = 3,067). The SNP rs887829 (UGT1A1) was significantly associated with TBIL levels (effect allele (T) frequency = 0.49, β (SE) = 0.59 (0.04), p = 9.13 × 10−54). Genome-wide conditional analysis and regional fine mapping pointed to rs887829 as a possible causal variant with a posterior inclusion probability of 0.99. The T allele of rs887829 is associated with lower hepatic expression of UGT1A1. Using rs887829 as an IV, two-stage least-squares MR showed a causal effect of bilirubin on hypertension (β = −0.76, 95% CI [−1.52, −0.01], p = 0.0459). Our finding confirms that UGT1A1 influences bilirubin levels. Notably, lower TBIL is causally associated with the increased risk of hypertension.
Similar content being viewed by others
Introduction
Hyperbilirubinemia has several causes, including hemolysis, cirrhosis, and bile duct obstruction. In the absence of liver disease, high circulating bilirubin levels have been associated with the reduced risk of several diseases, including respiratory diseases1; oxidative stress-mediated diseases, such as diabetes mellitus, diabetic nephropathy, cancer, and cardiovascular disease2,3,4,5,6,7,8; and hypertension9. The protective effect of bilirubin may be due to antioxidant activity10,11,12,13. There is also evidence that higher bilirubin is associated with the better survival and functional independence in the elderly14,15.
The heritability of serum bilirubin has been estimated to be 48 ± 6% (ref. 16). The genetic architecture of circulating bilirubin levels has been studied through genome-wide association studies (GWAS), which have revealed several associated variants in one locus, UGT1A1 (refs. 17,18,19). UGT1A1 has been found to be a major locus for serum total bilirubin (TBIL) in studies of Han Chinese20, individuals with European ancestry21,22, and African Americans23. The SNP rs887829 in UGT1A1 accounts for 12% of the variance of TBIL levels in African Americans23. UGT1A1, a UDP-glucuronosyltransferase, is the sole enzyme that glucuronidates bilirubin.
Mendelian randomization (MR) is a technique for using genetic variants as instrumental variables (IVs) to estimate the casual effect between exposure and outcome, and has been successfully used in studies of cardiovascular diseases, type 2 diabetes (T2D), heart failure, stroke, and nephrology19,24,25,26,27,28,29,30. The genetic architecture of common, complex diseases generally consists of common genetic variants with small effect sizes, and it is typically not possible to draw inferences using MR with only a single variant26. However, serum metabolites may have simpler genetic architectures31, sometimes with a single variant that can be used as a strong IV by itself27,28,29,30.
Here, we describe a GWAS for serum TBIL in West Africans. We confirm that UGT1A1 is a major locus for serum TBIL, with conditional analysis revealing only one association signal indexed by rs887829. Using rs887829 as an IV, we demonstrate a causal effect of serum TBIL on hypertension in African Americans, with lower serum (TBIL) being a causal risk factor for hypertension.
Results
Population description and genetic architecture
The characteristics of participants in the GWAS and MR studies are presented in Table 1 and Supplementary Table 1, respectively. In all studies, men had a higher mean serum TBIL level than women. Also, in all studies, mean age and prevalence of hypertension was similar for both men and women; however, men had a lower mean body mass index (BMI) compared to women. Overall, the mean age and prevalence of hypertension were higher, and mean serum (TBIL) levels were lower in the sampled West Africans than in the African Americans.
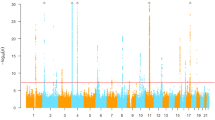
Heritability for serum TBIL in West Africans was estimated as 0.385 (SD 0.075). Only one locus, centered around UGT1A1 on chromosome 2, was genome-wide significant (Fig. 1), with the index SNP being rs887829 (T allele, effect allele frequency = 0.49, β = 0.59, p = 9.13 × 10−54, Supplementary Table 2). There was no inflation of the test statistic as assessed by a genomic control inflation factor (\(\lambda _{\mathrm{GC}}\)) of ~1.01 (Supplementary Fig. 1). The UGT1A1 locus included 205 SNPs (Supplementary Table 2) spanning 419 kb (Fig. 2). Among variants with a minor allele frequency (MAF) of at least 1%, Bayesian fine mapping of the UGT1A1 locus identified one causal variant, rs887829, with a marginal posterior inclusion probability of 0.99 (Fig. 2). Conditional on rs887829, there was no other genome-wide significant signal (Supplementary Fig. 2).
The x-axis represents position in Mb and the y-axis represents −log10 p values. Sky-blue lines represent recombination rates (cM/Mb) from the 1000 Genomes Project. LD blocks are shown using horizontal lines: red lines for Africans (AADM), orange for African Americans (HUFS), black for CEU from the 1000 Genomes Project, and blue for CHB from the 1000 Genomes Project. The red dot represents rs887829 and the blue dot represents rs10929302. The pink vertical line represents the position of rs3064744 (not present in these data). (Bottom) posterior inclusion probabilities based on fine mapping. The red vertical line represents the posterior inclusion probability of rs887829.
We next sought to confirm the association of rs887829 with serum TBIL using previously reported GWAS findings in the NHGRI-EBI GWAS Catalog17,18. Most previous studies (45/56 or ~80%) identified significant loci in a haplotype block containing rs887829, based on linkage disequilibrium patterns in European ancestry (CEU) and East Asian ancestry (CHB) individuals (Fig. 2 and Supplementary Table 3). This haplotype block was 5.68 kb in West Africans and 5.86 kb in African Americans, compared to 41.80 kb in CEU and CHB.
UGT1A1 is the sole enzyme responsible for the glucuronidation of bilirubin, which occurs in the liver and is necessary for making bilirubin water soluble. We reasoned that if UGT1A1 functions to make bilirubin excretable and hence to lower levels of bilirubin in serum, then the T allele of rs887829 should be associated with lower hepatic expression of UGT1A1. To test this hypothesis, we interrogated the GTEx database. Consistent with our hypothesis, the T allele of rs887829 was associated with lower expression of UGT1A1 in liver (p = 6.3 × 10−8).
Additive Bayesian network
The relationships among rs887829, serum TBIL, hypertension status, sex, age, BMI, and T2D status were evaluated using an additive Bayesian network (ABN) in individuals with African ancestry. Heuristic searching demonstrated that hypertension status was directly influenced by serum TBIL and indirectly influenced by the genetic risk score via serum bilirubin levels (Fig. 3).
Mendelian randomization
We then performed MR analysis in African Americans to assess the causal effect of serum TBIL on hypertension. To determine whether the MR approach was appropriate, we conducted preliminary analyses to evaluate the following key assumptions. First, there should be a strong association between the IV and the exposure. An F-st atistic is used to measure of the strength of the IV, with a threshold of the F-statistic > 10 indicating sufficient strength32. F-statistics of 332.83 and 193.89 were obtained for rs887829 in HUFS and CARDIA, respectively, indicating that rs887829 by itself is a strong IV for serum TBIL. Second, there should be no association between the IV and the outcome, conditional on the exposure. To evaluate this assumption, we regressed hypertension on rs887829, adjusting for serum TBIL as well as age, sex, BMI, and PCs. We observed no effect of rs887829 on hypertension beyond the effect mediated by serum TBIL in HUFS [OR (95% CI) = 1.01 (0.70, 1.73), and p = 0.6853] or CARDIA [OR (95% CI) = 1.21 (0.89,1.66), p = 0.2294). A third key assumption is that there should be no unmeasured confounders of the association between the IV and the outcome. Although we cannot test for the presence of unmeasured confounders, we included several measured covariates as possible confounders in the regression of prevalent hypertension on rs887829. Finally, we found that higher serum TBIL confers significant protection against prevalent hypertension, with β (95% CI) = −0.76 (−1.52, −0.01), and p = 0.0459 in combined analysis of African Americans (Fig. 4).
We estimated the conditional power of the MR analysis based on the African American data. Based on a combined sample size n = 3067, a significance level α = 0.05, a prevalence of hypertension of 0.26, a 0.46 odds ratio of hypertension per standard deviation of serum TBIL, and 12% of the variance of the exposure explained by the IV in African Americans23, we estimated a conditional power of 0.99.
Discussion
UGT1A1 is known to be a major locus influencing bilirubin levels. In the present study, we confirmed this observation in a study of West Africans and extended the original observation. Taking advantage of the shorter haplotypes and weaker LD in West Africans, we refined the significant region from 10 kb in individuals with European ancestry33 to <6 kb.
Previous studies have indicated the functionality of rs887829 or a very close proxy. For example, in a study of infants, the variability of TBIL levels explained by rs887829 increased from 7.0% (day 6) to 10.2% (day 7), consistent with the development of UGT1A1 isoenzyme expression34. The polymorphism rs3064744 is a (TA)n tandem repeat covering the TATA box of UGT1A1 and is a close proxy of rs887829 (r2 = 0.99)1,33,35,36; however, rs3064744 was neither genotyped nor imputed in our study. The SNP rs10929302 (3 kb upstream of rs3064744) is in a phenobarbital response enhancer module and is associated with the response to the anticancer drug irinotecan37. Thus, a pleiotropic region (~3 kb, from rs10929302 to rs34983651) influences serum bilirubin levels, phenobarbital induction of UGT1A1 expression, irinotecan response, and xenobiotic metabolism23,38.
The results of ABN modeling suggest that the effect of rs887829 is indirectly associated with the hypertension via an effect mediated by bilirubin. This relationship has been demonstrated experimentally in animal models. Mice treated with indinavir, which induces moderate hyperbilirubinemia by targeting hepatic UGT1A1, had reduced development of hypertension upon Ang II administration compared to mice that did not receive indinavir39. The effect was also observed in mice with hyperbilirubinemia, as a result of direct intravenous infusion of bilirubin39.
In the present study, we used MR to determine whether serum TBIL causally affects hypertension. Using rs887829 as an IV in a two-stage model, we found evidence of a causal role of serum TBIL on the prevalence of hypertension in African Americans, with lower serum TBIL associated with the increased risk of hypertension. The proportion of variance of the exposure explained by the IVs has a strong effect on the power of MR analysis. One genetic variant, rs887829, explained 12% of the variance of the exposure23, so the MR analysis, although based on a single IV, was well powered. In addition, our GWAS was performed on Africans from Ghana and Nigeria, whereas our MR analysis was performed on African Americans. We previously showed that African Americans share ~80% ancestry with West Central Africans, such as Yoruba from Nigeria40. To adjust for genetic ancestry, we included principal components in both the GWAS and MR analyses.
The MR analysis indicated that higher serum TBIL is causally associated with a lower risk of hypertension. In mice, bilirubin is a selective ligand for PPARα, driving the expression of genes that result in reduced white adipose tissue size, an increased number of mitochondria, reduced fat accumulation, and reduced insulin resistance41. In rats, stimulation of PPARα results in a lowering of blood pressure through increased expression of SOD-1, eNOS, and angiotensin II receptors, implicating protection against hypertension through mechanisms involving antioxidants and nitric oxide42. Taken together, these findings suggest that PPARα may be mediating the protective effects of higher serum TBIL on a range of cardiovascular and metabolic diseases.
In summary, rs887829, or a variant in strong LD with it, is likely a causal variant for serum TBIL, possibly via expression of UGT1A1 in the liver. An association between rs887829 and hypertension is mediated through serum TBIL. In African Americans, lower serum TBIL is causally associated with the risk of hypertension.
Methods
Genome-wide association analysis
West African individuals were drawn from the Africa America Diabetes Mellitus (AADM) study43,44,45,46. We included participants recruited from Ghana or Nigeria. Weight was measured in light clothes on an electronic scale to the nearest 0.1 kg and height was measured with a stadiometer to the nearest 0.1 cm. BMI was computed as weight (kg) divided by the square of height in meters (m2). The definition of T2D was based on the American Diabetes Association criteria. Blood samples were drawn after an overnight fast of at least 8 h. Serum TBIL was measured on a COBAS Integra 400 Plus (Roche Diagnostics, Indianapolis, IN, USA) using the Diazo method for a total of 1127 individuals. Participants in the AADM study were genotyped on high-density GWAS arrays (either the Affymetrix® Axiom® Genome-Wide PanAFR Array Set or the Illumina Multi-Ethnic Genotyping Array). After exclusions based on technical quality control (individual call rate ≤ 95%, SNP call rate ≤ 95%, Hardy–Weinberg equilibrium p < 10−6, and MAF < 0.01), imputation was performed using the African Genome Resources Panel available from the Sanger Imputation Service (https://imputation.sanger.ac.uk/). An info score ≥ 0.3 and MAF ≥ 0.01 were used for filtering imputed variants. Serum TBIL values were regressed on age and sex. The resulting residuals were ranked and inverse normalized. Association analysis was performed using the EPACTS (Efficient and Parallelizable Association Container Toolbox) pipeline (http://genome.sph.umich.edu/wiki/EPACTS), with BMI, T2D status, and significant principal components obtained from the R package SNPRelate47 as fixed effects, and genetic cryptic relatedness matrix as a random effect. Based on the Tracy Widom test, three principal components were significant48.
Fine mapping
Fine mapping was performed using the R package CAVIARBF, an approximate Bayesian method that can incorporate functional annotation49. Minimal data requirements are marginal statistical test results and linkage disequilibrium between SNPs. SNP annotations were coded for the absence (0) or presence (1) of promoter histone marks, enhancer histone marks, DNAse I hypersensitive sites, or bound proteins as provided by HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). Bayes factors were calculated conditional on a maximum number of causal SNPs. The estimated Bayes factors and prior probabilities were then used to estimate the posterior inclusion probabilities.
SNP-based heritability
We estimated the heritability of serum TBIL using LDAK50,51. A total of 506,737 genotyped SNPs with MAF > 0.05 were used. Log-transformed serum bilirubin levels and covariates of age, sex, BMI, and T2D were used for this estimation. First, based on local LD, we calculated a weight for each predictor (SNP), which showed how well each SNP was tagged. Next, using these weights, we calculated a kinship matrix to improve poorly tagged predictors that had lower than average MAF. Then, we fit the linear mixed model \({\mathrm{Var}}\left( {\mathbf{Y}} \right) = \sigma _g^2{\mathbf{G}} + \sigma _e^2{\mathbf{I}}\), in which Y is the vector of phenotype values, G is a kinship matrix based on the weighted SNPs, and I is an identity matrix. The estimates \(\hat \sigma _g^2\) and \(\hat \sigma _e^2\) were obtained via restricted maximum likelihood51. The proportion of phenotypic variance explained by additive genetic effects at common genotyped variants was estimated as \(\hat h_{\mathrm{snp}}^2 = \frac{{\hat \sigma _g^2}}{{\hat \sigma _g^2 + \hat \sigma _e^2}}.\)
Additive Bayesian network modeling
An ABN was used to evaluate relationships among rs887829, hypertension status, and serum TBIL. An ABN is a probabilistic graphic model52, extends to the generalized linear model (GLM), and reveals interdependencies among factors which may not be discovered in GLM. In contrast to standard multivariable regression analysis, which only shows interactions between risk factors and the outcome, an ABN illustrates the interactions between all variables. ABN modeling comprises three interrelated parts: parameter learning, network scoring, and structure learning53, and were implemented in the R package abn. The standard local heuristic search54 was applied to seven nodes (rs887829, bilirubin, hypertension, sex, age, BMI, and T2D) to evaluate node relationships. The majority consensus network was constructed from all edges present in at least 50% of the locally optimal networks found across 20,000 heuristic searches.
Mendelian randomization analysis
MR was performed to investigate whether serum TBIL has a causal effect on hypertension. Let Z represent the genetic variants used as IVs, let X represent serum TBIL as the exposure, and let Y represent hypertension status as the outcome. Let \(\hat X\) represent the values of X predicted by Z. The causal estimate of the exposure (\(\hat X\)) on outcome (Y), \(\beta _{\hat XY}\), can be estimated through two-stage least-squares regression. First, the exposure \(\hat X\) is estimated by calculating the fitted values from the regression of X on Z. Second, \(\beta _{\hat XY}\) is obtained by regressing Y on \(\hat X\). In the data analysis, we first regressed serum TBIL on genetic variants (rs887829, C allele as reference allele) and covariates sex, age, BMI, and PCs to estimate \(\hat X\). Second, we regressed hypertension (Y) status on \(\hat X\) to estimate \(\beta _{\hat XY}\), adjusting for sex, age, BMI, and PCs. Analysis was performed using SAS 9.4 (SAS Institute Inc. Cary, NC, USA). Two studies of African Americans, the Howard University Family Study (HUFS, n = 1933), and the Coronary Artery Risk Development in Young Adults (CARDIA) study (n = 1134), were used for MR analysis. Study procedures in HUFS, including serum TBIL assays and genotyping, have been described previously23,55. Participants were not ascertained based on any phenotype. Data from the CARDIA study were retrieved from dbGaP under study accession number phs000285.v3.p2. Meta-analysis with inverse variance weights was performed using the R package meta56. Power calculations were performed using mRnd (https://shiny.cnsgenomics.com/mRnd/)57.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The AADM and HUFS datasets used and/or analyzed in the current study are available from the corresponding author upon reasonable request for collaborative studies that are consistent with the IRB approvals and patient consent. The CARDIA data (phs000285) are available through dbGaP authorized approval.
Code availability
CAVIARBF is fully available on bitbucket (https://bitbucket.org/Wenan/caviarbf). The bitbucket page describes all dependencies and versions. HaploReg is a fully available on broadinstitute (https://pubs.broadinstitute.org). The broadinstitute web page describes all dependencies and versions. LDAK is a fully available on dougspeed (http://dougspeed.com). The dougspeed web page describes all dependencies and versions. Additive Bayesian network modeling in R is a fully available on http://r-bayesian-networks.org. The web page describes all dependencies and versions. A R package of meta is available on cran (https://cran.r-project.org). The cran web page describes all dependencies and versions. A R package of SNPRelate is available on cran (https://cran.r-project.org). The cran web page describes all dependencies and versions. mRnd is fully available on https://shiny.cnsgenomics.com/mRnd/. The web page describes all dependencies and versions.
References
Horsfall, L. J. et al. Serum bilirubin and risk of respiratory disease and death. JAMA 305, 691–697 (2011).
Novotny, L. & Vitek, L. Inverse relationship between serum bilirubin and atherosclerosis in men: a meta-analysis of published studies. Exp. Biol. Med. 228, 568–571 (2003).
Schwertner, H. A. & Vitek, L. Gilbert syndrome, UGT1A1*28 allele, and cardiovascular disease risk: possible protective effects and therapeutic applications of bilirubin. Atherosclerosis 198, 1–11 (2008).
Hunt, S. C. et al. Association of plasma bilirubin with coronary heart disease and segregation of bilirubin as a major gene trait: the NHLBI family heart study. Atherosclerosis 154, 747–754 (2001).
Breimer, L. H., Wannamethee, G., Ebrahim, S. & Shaper, A. G. Serum bilirubin and risk of ischemic heart disease in middle-aged British men. Clin. Chem. 41, 1504–1508 (1995).
Lin, J. P., Vitek, L. & Schwertner, H. A. Serum bilirubin and genes controlling bilirubin concentrations as biomarkers for cardiovascular disease. Clin. Chem. 56, 1535–1543 (2010).
Han, S. S. et al. High serum bilirubin is associated with the reduced risk of diabetes mellitus and diabetic nephropathy. Tohoku J. Exp. Med. 221, 133–140 (2010).
Horsfall, L. J., Burgess, S., Hall, I. & Nazareth, I. Genetically raised serum bilirubin levels and lung cancer: a cohort study and Mendelian randomisation using UK Biobank. Thorax 75, 955–964 (2020).
Wang, L. & Bautista, L. E. Serum bilirubin and the risk of hypertension. Int. J. Epidemiol. 44, 142–152 (2015).
Ziberna, L., Martelanc, M., Franko, M. & Passamonti, S. Bilirubin is an endogenous antioxidant in human vascular endothelial cells. Sci. Rep. 6, 29240 (2016).
Sedlak, T. W. et al. Bilirubin and glutathione have complementary antioxidant and cytoprotective roles. Proc. Natl Acad. Sci. USA 106, 5171–5176 (2009).
Jansen, T. & Daiber, A. Direct antioxidant properties of bilirubin and biliverdin. is there a role for biliverdin reductase? Front. Pharm. 3, 30 (2012).
DiNicolantonio, J. J., McCarty, M. F. & O’Keefe, J. H. Antioxidant bilirubin works in multiple ways to reduce risk for obesity and its health complications. Open Heart 5, e000914 (2018).
Sedlak, T. W. & Snyder, S. H. Bilirubin benefits: cellular protection by a biliverdin reductase antioxidant cycle. Pediatrics 113, 1776–1782 (2004).
Han, S. S. et al. Survival benefit of high serum bilirubin and low alkaline phosphatase in older adults. J. Am. Geriatr. Soc. 58, 1413–1415 (2010).
Lin, J. P., Cupples, L. A., Wilson, P. W., Heard-Costa, N. & O’Donnell, C. J. Evidence for a gene influencing serum bilirubin on chromosome 2q telomere: a genomewide scan in the Framingham study. Am. J. Hum. Genet. 72, 1029–1034 (2003).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Shah, S. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 11, 163 (2020). ARTN.
Lin, R. et al. Common variants of four bilirubin metabolism genes and their association with serum bilirubin and coronary artery disease in Chinese Han population. Pharmacogenet. Genom. 19, 310–318 (2009).
Johnson, A. D. et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 18, 2700–2710 (2009).
Sanna, S. et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 18, 2711–2718 (2009).
Chen, G. et al. UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur. J. Hum. Genet. 20, 463–468 (2012).
Lanktree, M. B., Theriault, S., Walsh, M. & Pare, G. HDL cholesterol, LDL cholesterol, and triglycerides as risk factors for CKD: a Mendelian randomization study. Am. J. Kidney Dis. 71, 166–172 (2018).
Pattaro, C. Genome-wide association studies of albuminuria: towards genetic stratification in diabetes? J. Nephrol. 31, 475–487 (2018).
Teumer, A. Common methods for performing Mendelian randomization. Front. Cardiovasc. Med. 5, 51 (2018).
Lee, S. J. et al. Bilirubin and stroke risk using a Mendelian randomization design. Stroke 48, 1154–1160 (2017).
Abbasi, A. et al. Bilirubin as a potential causal factor in type 2 diabetes risk: a Mendelian randomization study. Diabetes 64, 1459–1469 (2015).
McArdle, P. F. et al. Association between bilirubin and cardiovascular disease risk factors: using Mendelian randomization to assess causal inference. BMC Cardiovasc. Disord. 12, 16 (2012). ARTN.
Stender, S., Frikke-Schmidt, R., Nordestgaard, B. G., Grande, P. & Tybjaerg-Hansen, A. Genetically elevated bilirubin and risk of ischaemic heart disease: three Mendelian randomization studies and a meta-analysis. J. Intern. Med. 273, 59–68 (2013).
Yu, B. et al. Genetic determinants influencing human serum metabolome among African Americans. PLoS Genet. 10, e1004212 (2014).
Martens, E. P., Pestman, W. R., de Boer, A., Belitser, S. V. & Klungel, O. H. Instrumental variables application and limitations. Epidemiology 17, 260–267 (2006).
Namjou, B. et al. A GWAS study on liver function test using eMERGE network participants. PLoS ONE 10, e0138677 (2015).
Kawade, N. & Onishi, S. The prenatal and postnatal-development of Udp-glucuronyltransferase activity towards bilirubin and the effect of premature birth on this activity in the human-liver. Biochem. J. 196, 257–260 (1981).
Sai, K. et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin. Pharm. Ther. 75, 501–515 (2004).
Zhou, Y. et al. Quantitative trait analysis of polymorphisms in two bilirubin metabolism enzymes to physiologic bilirubin levels in Chinese newborns. J. Pediatr. 165, 1154–1160 (2014).
Innocenti, F. et al. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J. Clin. Oncol. 27, 2604–2614 (2009).
Zhou, J., Tracy, T. S. & Remmel, R. P. Bilirubin glucuronidation revisited: proper assay conditions to estimate enzyme kinetics with recombinant UGT1A1. Drug Metab. Dispos. 38, 1907–1911 (2010).
Vera, T., Granger, J. P. & Stec, D. E. Inhibition of bilirubin metabolism induces moderate hyperbilirubinemia and attenuates ANG II-dependent hypertension in mice. Am. J. Physiol. Reg. Integr. Comp. Physiol. 297, R738–R743 (2009).
Shriner, D. et al. Multiple loci associated with renal function in African Americans. PLoS ONE 7, e45112 (2012).
Gordon, D. M. et al. Bilirubin remodels murine white adipose tissue by reshaping mitochondrial activity and the coregulator profile of peroxisome proliferator-activated receptor α. J. Biol. Chem. 295, 9804–9822 (2020).
Ibarra-Lara, L. et al. PPARα stimulation exerts a blood pressure lowering effect through different mechanisms in a time-dependent manner. Eur. J. Pharm. 627, 185–193 (2010).
Rotimi, C. N. et al. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann. Epidemiol. 11, 51–58 (2001).
Adeyemo, A. A. et al. Evaluation of genome wide association study associated type 2 diabetes susceptibility loci in sub Saharan Africans. Front. Genet. 6, 335 (2015).
Adeyemo, A. A. et al. ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Nat. Commun. 10, 3195 (2019). ARTN.
Chen, G. et al. Refining genome-wide associated loci for serum uric acid in individuals with African ancestry. Hum. Mol. Genet. 29, 506–514 (2020).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Liu, Z. et al. Admixture mapping identifies genetic regions associated with blood pressure phenotypes in African Americans. PLoS ONE 15, e0232048 (2020).
Chen, W. et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200, 719–736 (2015).
Speed, D. & Balding, D. J. SumHer better estimates the SNP heritability of complex traits from summary statistics. Nat. Genet. 51, 277–284 (2019).
Speed, D., Hemani, G., Johnson, M. R. & Balding, D. J. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91, 1011–1021 (2012).
Korb, K. B. & Nicholson, A. E. Bayesian Artificial Intelligence (Chapman & Hall/CRC Press, 2004).
Lewis, F. I., Brulisauer, F. & Gunn, G. J. Structure discovery in Bayesian networks: an analytical tool for analysing complex animal health data. Prev. Vet. Med 100, 109–115 (2011).
Heckerman, D., Geiger, D. & Chickering, D. M. Learning Bayesian networks - the combination of knowledge and statistical-data. Mach. Learn. 20, 197–243 (1995).
Adeyemo, A. et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 5, e1000564 (2009) .
Balduzzi, S., Rucker, G. & Schwarzer, G. How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health 22, 153–160 (2019).
Brion, M. J. A., Shakhbazov, K. & Visscher, P. M. Calculating statistical power in Mendelian randomization studies. Int J. Epidemiol. 42, 1497–1501 (2013).
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health. We are thankful to the participants of the AADM project, their families, and their physicians. The study was supported in part by the Intramural Research Program of the National Institutes of Health in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, and the Office of the Director at the National Institutes of Health (1ZIAHG200362). Support for participant recruitment and initial genetic studies of the parent AADM study was provided by NIH grant 3T37TW00041-03S2 from the Office of Research on Minority Health. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 02/19/2020. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The data from CARDIA used for the analyses in this manuscript were obtained from dbGaP through accession study number phs000285.
Author information
Authors and Affiliations
Contributions
G.C. and D.S. analyzed the data and drafted the manuscript; A.A. and C.N.R. designed the study, interpreted the results, and revised the manuscript, J.Z. managed the data, and all authors read and apporoved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, G., Adeyemo, A., Zhou, J. et al. A UGT1A1 variant is associated with serum total bilirubin levels, which are causal for hypertension in African-ancestry individuals. npj Genom. Med. 6, 44 (2021). https://doi.org/10.1038/s41525-021-00208-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-021-00208-6