Abstract
PIK3CA mutations are the most common in breast cancer, particularly in the estrogen receptor-positive cohort, but the benefit of PI3K inhibitors has had limited success compared with approaches targeting other less common mutations. We found a frequent allelic expression imbalance between the missense mutant and wild-type PIK3CA alleles in breast tumors from the METABRIC (70.2%) and the TCGA (60.1%) projects. When considering the mechanisms controlling allelic expression, 27.7% and 11.8% of tumors showed imbalance due to regulatory variants in cis, in the two studies respectively. Furthermore, preferential expression of the mutant allele due to cis-regulatory variation is associated with poor prognosis in the METABRIC tumors (P = 0.031). Interestingly, ER−, PR−, and HER2+ tumors showed significant preferential expression of the mutated allele in both datasets. Our work provides compelling evidence to support the clinical utility of PIK3CA allelic expression in breast cancer in identifying patients of poorer prognosis, and those with low expression of the mutated allele, who will unlikely benefit from PI3K inhibitors. Furthermore, our work proposes a model of differential regulation of a critical cancer-promoting gene in breast cancer.
Similar content being viewed by others
Introduction
Activating oncogenic mutations are often characterized by gain-of-function single-base alterations or focal DNA copy-number amplification, where the gain of just a single copy of a mutant allele is sufficient for tumorigenesis1. These gains change the stoichiometric balance between mutant and wild-type alleles and are selected for in cancers, affecting approximately half of all oncogenic driver mutations1. Ultimately, they could dictate prognosis and therapeutic sensitivity.
However, the impact of gene dosage differences of oncogenic mutations generated at the gene expression level has been largely unexplored. Genetic variation and mutations regulate gene expression in an allele-specific manner—known as cis-regulatory variation2—by altering protein and miRNA binding, for example. Normal cis-regulatory variation affects most of the human genome in all tissues and generates the wealth of phenotypic variation seen in species3,4,5. Moreover, an extensive contribution from noncoding variants to RNA alterations was recently observed in tumors6, including allelic imbalance of somatic mutations7.
Nevertheless, one unsolved aspect is how much each mechanism contributes to generating allelic imbalances in expression and whether they do it independently or in synergy. In breast tissue, germline regulatory variation is associated with disease risk8 and affects frequently mutated genes9. We and others have shown that variants affecting the expression levels of BRCA1 and BRCA2 modify the risk of breast cancer in germline mutation carriers10,11. We found that carriers of germline nonsense mutations in the tumor suppressor gene BRCA2 were at a lower risk of developing breast cancer when the remaining wild-type allele was highly expressed11.
Here, we hypothesize that cis-regulatory variation also modulates the penetrance of oncogenic coding mutations. In the context of a gene cis-regulated by a genetic variant generating imbalanced allelic expression, we postulate that an oncogenic activating mutation in the same gene will have a different clinical impact depending upon whether it occurs in the preferentially expressed allele or the less expressed one. We tested this model in the context of heterozygous mutations in PIK3CA, the most frequently mutated gene in breast cancer. First, we investigated whether normal cis-regulatory variation regulated the expression of PIK3CA in normal breast tissue. Then, we calculated allelic expression ratios between mutant and wild-type copies in tumors from two large breast cancer datasets—METABRIC and TCGA—both normalized for DNA copy number or not. Finally, we correlated the allelic expression ratios with clinical data. This approach allows us to distinguish between expression imbalances generated from cis-regulatory variation alone, altered DNA copy number, or both mechanisms.
results
Normal cis-regulatory variation affects PIK3CA expression in healthy breast tissue
To investigate whether cis-regulatory variation modulates the expression of PIK3CA in normal breast tissue, we analyzed data from previous allelic expression analysis of normal breast tissue from 64 healthy donors12. We calculated the ratio of expression of one allele by the other in heterozygous variant positions, which is a robust approach to detect cis-acting variant effects, as it cancels out the trans effects that act on the same gene and influence both alleles equally. We found six variants in PIK3CA displaying differential allelic expression (daeSNPs) (see “Methods”) (Fig. 1). Of these six, only rs3729679 is not in strong linkage disequilibrium (LD) with the others (Supplementary Table 1). rs3960984 showed the largest proportion of heterozygotes displaying allelic differences (57%), while three other daeSNPs shared the smallest fraction (14%): rs12488074, rs4855093, and rs9838411.
AE ratios for six daeSNPs in the PIK3CA gene region, each dot is a heterozygous individual for the corresponding variant indicated in the x-axis, dotted lines delimit the levels of 1.5-fold difference for either allele preferential expression (\(\left|{{{\rm{AE}}}}\right|=0.58\)). Boxplots display the median, the lower and upper hinges corresponding to the first and third quartiles, and lower and upper whiskers corresponding to the smallest and largest values from the 1.5 * IQR (interquartile range), respectively.
In the daeSNPs rs7636454, rs3960984, rs12488074, and rs9838411, the ratios showed a unilateral distribution, with samples displaying preferential expression towards the same allele. These patterns of allelic expression ratios’ distribution suggest that the daeSNPs at which allelic expression is being measured and the possible functional regulatory variants (rSNPs) are in strong, yet incomplete, LD with each other13.
While the mapping analysis carried out to identify candidate rSNPs did not find a significant association after multiple testing correction (Supplementary Table 2), one of the variants with nominal P value ≤ 0.05, rs2699887 (Wilcoxon two-sample test estimated difference of 0.22, 95% CI = [0.031-Inf]) (Supplementary Fig. 1A), showed great regulatory potential. Namely, it is an eQTL (expression quantitative trait locus) for PIK3CA (P = 0.011, Supplementary Fig. 1B) in tumors from METABRIC14, is located at its promotor region and at a DNAse I hypersensitivity site (Supplementary Fig. 1C), and is bound by POL2 in a breast cancer cell line (Supplementary Table 3). In-silico functional analysis of this variant suggested a disruption of the binding motif of the transcription factor NF-YA (Supplementary Fig. 1D), and in vitro studies revealed a preferential protein::DNA binding to the minor T allele of rs2699887, which is associated with higher expression of PIK3CA in tumors (Supplementary Fig. 1E).
Preferential expression of the PIK3CA mutated alleles is frequent in breast tumors
Changes in DNA copy number in tumors are associated with changes in gene expression in cis1,14,15,16,17 leading to dosage imbalances of coding mutations7. However, these differences can also be due to germline and somatic cis-regulatory variation, but their effect on mutation dosage imbalance is underexplored. So, we set out to assess whether PIK3CA somatic mutations would have their functional effects, or penetrance, modified by imbalances in allelic expression generated by regulatory variants. We hypothesized that preferential expression of a gain-of-function mutation would have a more substantial clinical impact than those occurring in lowly expressed alleles, thus generating intertumor clinical heterogeneity (Fig. 2a). To test this, we carried mutant vs. wild-type allelic expression analysis in breast tumor samples carrying somatic PIK3CA missense mutations on two independent sets of data, the METABRIC (n = 94) and the TCGA (n = 178) projects. Supplementary Table 4 presents a summary description of the two datasets and Supplementary Fig. 2 shows the number, location, and amino acid alterations of the mutations across the two datasets.
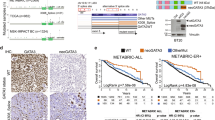
a Schematic representation of the hypothesis: cis-acting regulatory variants (rVar), either from germline or somatically acquired, generate different relative allelic expression ratios of mutant and wild-type alleles, resulting in tumors of different prognosis. b Top: log ratio α, β, and γ 89% credible intervals (CI) in breast tumors. Bottom: CIs collective posterior distribution split according to imbalance. A sample is deemed imbalanced if the CI does not cross zero. Samples with significant imbalance are displayed in red. c Correlation analysis of α vs. β and α vs. γ, showing that both genomic copy-number dosage and allelic expression regulation contribute to imbalances in the expression of mutated alleles in tumors. Point coordinates are Maximum A Posteriori probability estimates (MAP) of the 89% CIs. d Comparison of matched γ and β values, showing predominance of tumors with a preferential allelic expression of the mutated allele. Point coordinates are Maximum A Posteriori probability estimates (MAP) of the 89% CIs.
Next, we calculated three allelic ratios from DNA-seq and RNA-seq data for each mutation:
-
(1)
\(\alpha ={\log }_{2}\) (number of mutant RNA-seq reads/number of wild-type RNA-seq reads), i.e., the net mutant allele expression imbalance;
-
(2)
\(\beta ={\log }_{2}\) (number of mutant DNA-seq reads/number of wild-type DNA-seq reads), i.e., the mutant allele relative copy-number;
-
(3)
γ = α − β, i.e., the net mutant allele expression imbalance normalized for the DNA allelic copy-number imbalances, which corresponds to a putative mutant allele expression imbalance due to cis-regulation.
In this way, α reports on the net allelic expression imbalance, generated by different mechanisms including copy-number aberrations, cellularity differences, and cis-regulatory variation, while γ reports specifically on the contribution from cis-regulatory variation (rVar in Fig. 2a), including normal genetic variation, somatic noncoding mutations, and allelic epigenetic changes. Figure 2b displays the distributions of the different ratios.
We found that net mutant allele expression imbalances (α ratio) are frequent in breast tumors, at 70.2% in METABRIC (66 out of 94) and 60.1% in TCGA (107 out of 178). The same is true for γ ratios, at 27.7% for METABRIC (26 out of 94) and 11.8% for TCGA (21 out of 178), indicating that cis-regulatory effects acting on mutations are also frequent in breast tumors. In both sets, we found samples with striking net preferential allelic expression for the mutant allele (maximum 44.8-fold and 220-fold in METABRIC and TCGA, respectively), but not so for the preferential expression of the wild-type allele (fold differences of 5.4 and 29 in METABRIC and TCGA, respectively) (Fig. 2b). Similarly, the mutant allele’s most pronounced preferential expression trend was found for the γ ratio, 10- and 4.2-fold for METABRIC and TCGA, respectively, albeit with smaller fold differences between alleles.
Interestingly, we observed that within the samples with significant mutant allele expression imbalance due to cis-regulatory variation there was a significant prevalence of samples that preferentially expressed the mutated allele in both datasets (binomial test Prob. = 1, 89%−CI = [0.89, 1.00], P = 3 × 10−8 for METABRIC and Prob. = 0.90, 89%−CI = [0.73, 0.98], P = 2 × 10−4 for TCGA).
Cis-regulatory variants contribute significantly to imbalances in the expression of mutant alleles
Next, hypothesizing that both copy number and cis-regulatory variants are the major contributors to allelic expression, we set out to assess the contribution of each mechanism toward the net mutant allele expression imbalances detected in these tumors. First, we found positive correlations between net allelic expression and both copy number and cis-regulatory variation (Fig. 2c), albeit with an effect for the copy number over the double the size of that found for cis-regulatory variation (average Pearson correlation r2 = 0.80 and 0.34, respectively). Next, we considered the variance (Var) of the net allelic expression as the sum of the effects of both mechanisms, plus the covariance (Cov) accounting for predicted non-mutual exclusion of mechanisms acting on any given allele:
we calculated the contribution of cis-regulatory variation to the variance of net allelic expression as \(\left({{{\rm{Var}}}}(\gamma )+{{{\rm{Cov}}}}(\beta ,\gamma )\right)/{{{\rm{Var}}}}(\alpha )\). Here, we found that cis-regulatory variants explain 20.6% and 14.4% of the variability of net mutant allelic expression seen in METABRIC and TCGA, respectively (Supplementary Table 5).
Finally, assessing how the two mechanisms act simultaneously on each tumor, we found that the majority of samples (70.2% and 54.5% for the METABRIC and TCGA, respectively) had positive γ and negative β values (Fig. 2d), suggesting that although the mutant allele was in lower genomic quantity, it was nevertheless preferentially expressed compared to the wild-type allele. Interestingly, there were 10.6% and 11.2% samples with positive α and negative β values, in METABRIC and TCGA respectively. This shows that these tumors overexpress the mutant allele despite this allele being in lower copy number.
Only a minor fraction of samples displayed co-occurring preferential allelic expression and a higher allele copy number of the mutant allele (6.38% and 8.43% for the METABRIC and TCGA, respectively). These results were independent of the effect of tumor cellularity (Supplementary Fig. 3).
Preferential expression of mutant alleles by cis-regulatory variation associates with poor prognosis
To investigate the impact of differential cis-regulation of PIK3CA’s mutations on clinical outcome (overall and disease-specific survival), we performed univariate survival analysis with γ ratios categorized in three groups, based on the existence of imbalance and its direction, i.e. whether there was significant predominance of expression of the mutated allele γmut, of the wild-type allele γwt, or balanced allelic expression γbalanced. We uncovered that the group γmut had a poorer disease-specific survival rate (P = 0.031, Fig. 3a) than the γbalanced group for METABRIC. The median overall survival for the γmut group was 5.88 years and for the γbalanced group was 12.46 years (Supplementary Fig. 4B), whereas, in the disease-specific analysis, the mean survival of the γmut patients was 7.07 years, and 41% of patients died during the length of the follow-up, in comparison with 25.3% deaths in the γbalanced group (Fig. 3a).
a Kaplan–Meier curve of disease-specific survival showing the worse prognosis of patients with differential expression of the PIK3CA mutations (γmut group, shown in blue) compared to those expressing equimolar levels of mutation and wild-type alleles (γbalanced group, shown in red), in METABRIC. Shown below the graph are the numbers of patients at risk per group throughout time. b Preferential expression of the mutated allele is associated with ER-negative, PR-negative, and Her2-positive breast tumors. In all graphs, samples were colored according to the significance of the allelic expression imbalance. q values indicated correspond to the Wilcoxon rank-sum test with continuity correction, corrected for multiple testing using the Benjamini & Hochberg method. Survival plots indicate the 95%CI as colored shades. Boxplots display the median, the lower and upper hinges corresponding to the first and third quartiles, and lower and upper whiskers corresponding to the smallest and largest values from the 1.5 * IQR (interquartile range), respectively.
The categorized γ ratios were not significantly associated with overall survival in the multivariate analysis (Supplementary Fig. 5). However, some of the variables that are usually independent prognosis factors, such as PR and HER2 statuses, were not significantly associated with survival either in this analysis. In the TCGA set, there was a trend toward a worse disease-specific survival of those patients whose tumors preferentially express the mutated allele (Supplementary Fig. 6). However, due to the relatively shorter follow-up time of this dataset (median ~1 year) and the fact that tumors were mainly Luminal A (~61.2% of samples)18, the power to detect significant differences is smaller than that of METABRIC. Nevertheless, the joint analysis of the two datasets showed a significantly worse disease-specific survival of the αmut group of patients, with a concordant trend in the γmut group (Supplementary Fig. 7).
PIK3CA preferential mutant allele expression associates with clinicopathological variables
Next, we sought to investigate whether PIK3CA’s differential mutant allele was associated with known prognostic clinicopathological variables, namely hormone receptors (ER, PR) and HER2 amplification, which are directly and indirectly connected to gene expression regulation, respectively.
For both datasets, we observed that preferential mutant allele expression driven by cis-regulatory variation (γ) was associated with markers of worse prognosis, namely it was significantly higher in ER-negative tumors and PR-negative tumors, and in HER2-positive tumors only in METABRIC (Fig. 3b). When evaluating the contribution of cis-regulatory variation to this association, we also found that higher average γ values associated with lower PR expression (P = 0.040) and HER2-positive tumors (P = 0.025), but we did not find a significant association with ER expression (P = 0.129) (Supplementary Fig. 8).
Given these results, we took γ into consideration in the survival analysis within the expression subgroups of ER, PR, and HER2, but did not find significant differences in overall and disease-specific survival in METABRIC (Supplementary Fig. 4).
Considering other known prognostic variables, including tumor size, grade, and molecular subtypes (PAM5019 and IntClust14, we found a significant association between γ ratios and PAM50 subtypes only in METABRIC (q = 0.027) (Supplementary Table 6 and Supplementary Fig. 9).
Finally, we did not find an association between the candidate germline regulatory variant rs2699887 and γ or clinical outcome, suggesting germline variants are unlikely to be involved in the significant associations described above (data not shown).
However, supporting the involvement of somatic cis-regulatory variants instead, we found smaller fold changes and less samples with imbalances measured at common PIK3CA variants in normal-matched tissue data than those measured at mutations in tumor tissue (Supplementary Fig. 10).
Discussion
Our work reveals the role of cis-regulatory variation acting on PIK3CA somatic mutations as modifiers of mutation penetrance. We show for the first time that allelic expression imbalance between PIK3CA’s mutant and wild-type alleles is common and prognostic in breast cancer.
Particularly, preferential expression of the mutant allele is significantly more common than that of the wild-type allele, and considering that PIK3CA is an oncogene, one possibility is that positive selection could have a role in generating this difference, which should be further investigated. Furthermore, we also found that allelic imbalance in expression observed for the mutant alleles in the tumors was greater than that observed for single-nucleotide polymorphisms in the normal-matched tissue of patients. These findings support the hypothesis of somatic regulatory mutations involvement in generating the imbalances observed in the tumors. While genomic allelic imbalance remains the largest determinant of allelic expression dosage (showing the highest correlation with and contributing the most to the variability observed in net allelic expression), cis-regulatory variation is also significantly correlated with net allelic expression and explains ~16% of its variability across samples in these sets of tumors.
The analysis of RNA-seq data from two independent cohorts of tumor samples, the METABRIC and TCGA projects, strongly supports our findings.
Moreover, we show that preferential expression of the mutant allele due to cis-variation is associated with poor prognosis variables, such as ER-negative, PR-negative, and Her2-positive statuses20,21. In the METABRIC dataset, we also found that preferential expression of the mutant allele was associated with worse overall and disease-specific survival. The high stringency in calling imbalance and the focus on a specific type of mutation in one gene, limits this study in terms of the sample size analyzed, but on the other hand it provides the simplest scenario for testing our hypothesis. Interestingly, the joint analysis of the datasets revealed some level of association between disease-specific survival and the preferential expression of the mutant allele, both net and due to cis-regulation, reaffirming the clinical importance of the expression level of a mutation commonly associated with aggressive tumors. In addition, some tumors presented preferential expression of the wild-type allele of PIK3CA, suggesting that these mutations are lowly expressed and possibly passenger events.
Besides the potential use of our findings as a prognosis biomarker in the clinic, these results may also have therapeutic implications. Some of the major clinical challenges in cancer treatment are identifying biomarkers of prognosis and defining which patients will benefit from a given therapy. Particularly, it is crucial to identify patients unlikely to respond to specific therapies to prevent unnecessary drug cytotoxicity without any therapeutic benefits. Our results reveal the importance of considering allelic expression in somatic mutation screens in these two aspects of patient management. Despite the high frequency of PIK3CA mutations in breast cancers, the response to PI3K inhibitor therapy has been more challenging than expected, and the prognostic significance of detecting somatic PIK3CA mutations in breast tumors is unclear22. Relevant to this discussion, we have previously shown that the presence of PIK3CA mutations confer a poorer prognosis in patients with ER-positive breast cancer only when stratified into copy-number driven subgroups (IntClust 1+, 2+, 9+)23.
In this study, we provide new evidence for the prognostic significance of these mutations at the expression level in breast tumors. Particularly for tumors with significant preferential expression of the wild-type allele, this prognostic significance has a potential impact on therapy response and clinical management since one may hypothesize that little to no benefit would come from treatment in the cases not expressing the targetable mutation.
Further studies evaluating the allelic expression of mutant oncogenes in the tumors of patients enrolled in molecular-driven trials will clarify this impact.
More challenging is determining which cis-regulatory mechanisms are promoting allelic expression imbalances. Both inherited11,24 and acquired variants6,25,26,27 can affect gene expression in an allelic manner28,29.
Here we show that normal cis-regulatory variation regulates PIK3CA’s expression in normal breast tissue, with the possible contribution of rs2699887 as a regulatory variant. We also found that the heterozygotes for “rs2699887” were associated with higher expression of the PIK3CA gene compared to the common homozygotes. Although there is published data supporting the clinical association of rs2699887 with poor prognosis in other cancers30,31, linked to an increase in PI3K signaling, there is still some data supporting the opposite association32,33. We did not find an association between rs2699887 and survival, which opens the possibility for other mechanisms besides normal cis-regulatory variation to be considered as contributors to the preferential allelic expression in these tumors (data not shown).
Double PIK3CA mutations in the same allele are frequent in breast tumors34, and the impact of noncoding mutations in cancer is just starting to be explored6. So, a possibility is that the combination of noncoding and coding mutations in the same gene might be underlying the allelic expression imbalances we are detecting.
Further studies on allelic expression imbalances of activating mutations, and even inactivating ones, should further reveal the contribution of cis-regulatory mechanisms in tumor development and progression. Particularly interesting to determine is whether the coding mutation originates in an allele predisposed with higher expression, or whether a sequence of somatic events introduces the coding activating mutation and additional cis-regulatory noncoding mutations. The answers could have significant repercussions on our understanding of tumor evolution.
In summary, we show that differential expression between the mutant and wild-type alleles of PIK3CA is common in breast cancer and with a significant contribution from allele-specific cis-regulatory effects. We further show that mutant allele differential expression is associated with clinical parameters such as ER, PR, and HER2 statuses and is prognostically significant.
Collectively, our work establishes the prognostic relevance of allele-specific transcriptional regulation of PIK3CA somatic mutations. It also supports a shift in the mutation testing in patient management, where the level of expression of these mutations should be considered, besides the detection at the DNA level.
Methods
Subjects
Normal breast and tumor samples were obtained with the written informed consent from donors and appropriate approval from local ethical committees, with the detailed information described in the respective original publications: normal tissue9, METABRIC14, TCGA35.
Differential allelic expression analysis
DNA and total RNA from 64 samples of normal breast tissue were hybridized onto Illumina Exon510S-Duo arrays (humanexon510s-duo), and data were analyzed as described before12. In short, after sample filtering and normalization, variants with average RNA log2 allelic intensity values greater than 9.5 and heterozygous in five or more samples were kept for further analysis.
Allelic log ratios were calculated for RNA and DNA intensity data:
for alleles A and B.Next, variants that showed significant differences between the RNA log ratios between heterozygous (AB) and homozygous groups (AA and BB) (two-sample Student’s t test, P value < 0.05) were selected for differential allelic expression analysis.
Allelic expression (AE) ratios were normalized for allelic DNA content:
Differential allelic expression (DAE) at the sample level was defined as ∣AE ratio∣ ≥ 0.58 (1.5-fold or greater between alleles), based on previous studies using microarray data3,36. Variants with at least 10% and three heterozygous samples displaying DAE were further classified as daeSNPs.
Linkage disequilibrium (LD) between daeSNPs was evaluated using the genetic variant-centered annotation browser SNiPA37.
Genotype imputation analysis on normal breast tissue samples
Illumina Exon 510 Duo germline genotype data from the 64 samples that passed microarrays quality control, were filtered to keep variants with call rates ≥85%, minor allele frequency >0.01, and Hardy–Weinberg equilibrium with P > 1 × 10−5. Next, genotypes were imputed with MACH1.038 for all additional known variants on chromosome 3, using as reference panel the phased CEU panel haplotypes from the HapMap3 release (HapMap3 NCBI Build39, CEU panel —Utah residents with Northern and Western European ancestry), and the recommended two-step imputation process: model parameters (crossover and error rates) were estimated before imputation using all haplotypes from the study subjects and running 100 Hidden Markov Model (HMM) iterations; then genotypes were imputed using the model parameter estimates from the previous round. Imputation results were filtered based on an \({{{\rm{rq}}}}\,{{{\rm{score}}}}\ge 0.3\)38, a platform-specific measurement of variant imputation uncertainty.
Differential allelic expression (DAE) mapping analysis on normal breast tissue samples
Differential allelic expression mapping analysis was performed by stratifying AE ratios at each PIK3CA daeSNP according to the genotype at variants located within ±250 Kb.
A Mann–Whitney test was applied to test if the mean of the absolute AE ratios of the heterozygous samples was greater than those of the combined reference and alternative allele homozygous samples. Correction for multiple testing was performed using BH method (p.adjust, R stats 4.0.3 package40) and limiting the significance to q values ≤0.05.
Functional annotation of DAE mapping associated variants
Variants in LD with SNPs with DAE mapping nominal-p-value ≤0.05 were retrieved using the function get_ld_variants_by_window from the ensemblr R package (https://github.com/ramiromagno/ensemblr) using the 1000 GENOMES project data (phase_3) for the EUR population and an r2 > 0.95. These proxy SNPs were assessed for overlap with epigenetic marks derived from the Encyclopedia of DNA Elements (ENCODE) and NIH Roadmap Epigenomics projects, such as chromatin states (chromHMM) annotation, regions of DNase I hypersensitivity, transcription factor binding sites, and histone modifications of epigenetic markers (H3K4Me1, H3K4Me3, and H3K27Ac) (http://genome.ucsc.edu/ENCODE/) for normal human mammary epithelial cells (HMECs), human mammary fibroblasts (HMFs), BR.MYO (breast myoepithelial cells) and BR.H35 (breast vHMEC) and two breast cancer cell lines MCF-7 and T47D. We prioritized variants located on either active promoter or enhancer regions in mammary cell lines, and for which ChIP-Seq data indicated protein binding or position weight matrix (PWM) scores predicted differential protein binding for different alleles. Two publicly available tools, RegulomeDB and HaploReg v4.1, and the MotifBreakR Bioconductor package, were also used to evaluate those candidate functional variants39,41,42.
Electrophoretic mobility shift assay (EMSA)
MCF-7 (ER-positive) and HCC1954 (ER-negative) breast cancer cell lines were cultured in DMEM and RPMI culture media, respectively, supplemented with 10% FBS and 1% PS (penicillin and streptomycin). Nuclear protein extracts were prepared using the Thermo Scientific PierceTM NER kit, according to the manufacturer’s instructions. Oligonucleotide sequences corresponding to the C (common) and T (minor) alleles of rs2699887 (5’-AGCGTGAGTAGAGCGCGGA[C/T]TGGCCGGTAGCGGGTGCGGTG-3’) were labeled using the Thermo Scientific Pierce Biotin 3’ End DNA Labelling Kit, according to the manufacturer’s instructions. Oligonucleotides with known binding motifs for NF-YA43 and E2F144 were used in competition assays. Undiluted antibodies used for supershift competition assays were NF-YA (H-209) (Santa Cruz Biotechnology, SC-10779X) and HMGA1a/HMGA1b (Abcam, ab4078). EMSA experiments were performed using the Thermo Scientific LightShiftTM Chemiluminescent EMSA Kit, using the buffer and binding reaction conditions previously described8. Each EMSA was repeated at least twice for all combinations of cell extract and oligonucleotide, which were also tested in serial dilution amounts.
Breast tumor samples
The METABRIC dataset of tumor samples included 2433 samples from the METABRIC project14 with DNA sequencing data, among which 480 were subjected to a capture-based RNA sequencing study23. Sequencing libraries were generated as previously described. In brief, sequencing libraries using total RNA generated from frozen tissues with a TruSeq mRNA Library Preparation Kit using poly-A-enriched RNA (Illumina, San Diego, CA, USA) and enriched with the human kinome DNA capture baits (Agilent Technologies, Santa Clara, CA, USA). Six libraries were pooled for each capture reaction, with 100 ng of each library, and sequenced (paired-end 51bp) on an Illumina HiSeq2000 platform. We selected a subset of samples with DNA and RNA sequencing data and PIK3CA missense mutations for further analysis.
The TCGA dataset comprised 695 samples from TCGA breast cancers35, from which we selected a subset of 289 samples with PIK3CA missense mutations for further analysis. Supplementary Table 3 summarizes the demographic features and disease characteristics of the two datasets.
DNA-seq and RNA-seq variants calling in tumors
Alignment and preprocessing
Sequence data (FASTQ) mapped to the reference genome (hg19) were aligned using STAR v2.4.145. A two-pass alignment was carried out: splice junctions detected in the first alignment run are used to guide the final alignment. Duplicates were marked with Picard v1.131 (http://picard.sourceforge.net). Genome Analysis Toolkit (GATK) was used for indel realignment and base quality score recalibration46.
Variant calling and annotations
SNV and indel variants were called using GATK Haplotype Caller. Hard filters using GATK VariantFiltration were applied to variants46. Variants were annotated with Ensembl Variant Effect Predictor (VEP)47. Heterozygous genotypes were called from DNA data to avoid RNA editing and other RNA-related variants because true allelic imbalance can lead to heterozygous sites being called homozygous in RNA-based genotype calling.
Analysis of allelic expression imbalances in tumors
Before the analysis, a set of filtering steps was performed to select samples: (1) presence of missense mutations; (2) and a minimum of 30 reads for RNA-seq and DNA-seq data48,49,50.
Clinical data for METABRIC were updated from the original studies with the latest available records. Clinical data for TCGA were imported from https://portal.gdc.cancer.gov/ on November 26, 2018.
Filtering of tumor samples
For both datasets —METABRIC and TCGA—, a set of quality control criteria were applied to filter the DNA-seq and RNA-seq samples, namely:
-
a.
Keep only samples containing PIK3CA missense mutations;
-
b.
Keep samples whose coverage at mutated loci is, all together in both alleles, at least 30 reads for both RNA-seq and DNA-seq data48,49,50.
Clinical data of METABRIC patients were updated from the original studies with the latest available records. The TCGA clinical dataset was obtained from cBioPortal51,52 on 28 November 2021 by programmatic access with the R package cgdsr.
Allelic expression imbalances in tumor data
Allelic expression imbalances are calculated as follows. For each mutated loci, the pair of read counts (X, Y), for wild-type (X) and mutant (Y) alleles, respectively, measured either by DNA-seq or RNAseq, are transformed using the log ratios β, α, and γ, which are defined as follows:
the DNA mutant allele ratio, which served to control for sequencing artifacts from heterozygous genotypes and to account for differences in variant frequencies in DNA;
that served as a measure of the net allelic expression imbalance in tumors;
the normalized mutant allele expression ratio, a proxy for the mutant allelic expression imbalance due to cis-regulation alone.
Statistical inference of allelic expression imbalances
According to these log-ratio definitions, a positive value indicates an imbalance toward the mutant allele, and a negative value an imbalance favoring the wild-type allele. However, the statistical significance of each log ratio depends on the read coverage of each allele, e.g., low read-coverage values are subject to greater random variation, and hence less reliable log ratios and imbalances estimation. To assign a measuring of uncertainty to our imbalances’ estimates, we assumed that the read counts are well modeled by a Beta-Binomial distribution, and following Bayesian reasoning, we estimated 89% credible intervals (CI) and Maximum A Posteriori probability estimates (MAP) for the log ratios β, α, and γ (reported in Fig. 2).
Allelic expression imbalances in normal-matched tissue data
Solid normal breast tissue from breast cancer female patients was obtained from TCGA-BRCA. We selected 112 samples with RNA-Seq data, obtained in bam file format. Sequence data were converted to fastq format (samtools53), underwent initial quality control (FastQC54), and trimming (Trimmomatic55). Following QC, six samples were removed from analysis. The remaining sequence data was mapped to the reference genome (hg38) using STAR aligner (v.2.7.7a45). Otherwise, alignment, preprocessing and variant calling was performed as described below. RNA data was filtered to contain only heterozygous variants at the DNA level, circumscribed to PIK3CA’s genomic location. DNA data was accessed from TCGA-BRCA’s microarray raw data for 111 of the 112 initial RNA-Seq samples. Genotypes were obtained using the CRLMM algorithm (‘crlmm’ R Bioconductor package,56) and quality controlled for HWE, major allele frequency and 10% missing genotypes (‘SNPassoc’ R package57). Genotypes were lifted over from hg38 to hg37 (‘rtracklayer’ R Bioconductor package58), harmonized (‘GenotypeHarmonizer’59), and imputed (Michigan Imputation Server60). Obtained genotypes were quality controlled using PLINK61 and lifted over back to hg38. Allelic expression imbalances, equivalent to α ratios in tumors, were inferred for heterozygous germline variants as described above.
Two-sample tests of imbalance ratios with clinical covariates
Association between allelic expression imbalance ratios and clinical data was achieved by bivariate analysis Wilcoxon rank-sum test with continuity correction or Kruskal–Wallis rank-sum test, as indicated in tables and figures. P values were adjusted per study using the Benjamini & Hochberg correction and were considered significant when ≤ 0.05.
Correlation analysis
Correlation analysis α vs β and α vs γ ratios for both sets of samples were performed using a Pearson’s test. All statistical analysis and data visualization were performed using R.
Survival analyses
Kaplan–Meier plots and multivariate Cox proportional hazard models were used to examine the association between alpha and gamma allelic expression ratios and survival using the survival package from R62,63. Death due to all causes was used as the endpoint, and all alive subjects were censored at the date of the last contact. Kaplan–Meier survival curves were compared using the log-rank test.
For the multivariate analysis, Cox proportional hazard model was used to assess the effect of γ on the overall survival. Hazard ratios (HRs) and 95% confidence intervals (CI) were estimated by fitting the Cox model while adjusting for age and tumor characteristics, such as size, Scarff–Bloom–Richardson histological grade, clinical stage and estrogen receptor (ER), progesterone (PR), and human epidermal growth factor 2 (HER2) statuses.
For the bivariate analysis, Wilcoxon rank-sum two-sample tests were used to compare α and γ between different hormone receptor statuses and q ≤ 0.05, calculated using the Benjamini & Hochberg method, were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Microarray raw data are deposited in the Gene Expression Omnibus under accession number GSE35023. Primary data (BAM files) for DNA-seq are deposited at the European Genome-phenome Archive (EGA) under study accession number EGAS00001001753 and may be downloaded upon request and authorization by the METABRIC Data Access Committee. Primary data (BAM files) for RNAseq are available from the authors upon reasonable request. Primary data (BAM files) for DNA-seq and RNAseq from TCGA are deposited in the database of Genotypes and Phenotypes (dbGaP) under the study accession number phs000178.
Code availability
The filtered data and code for the analysis of mutant allele expression imbalances and the survival analysis can be publicly accessed at https://github.com/maialab/npjbcPIK3CA.
References
Bielski, C. M. et al. Widespread selection for oncogenic mutant allele imbalance in cancer. Cancer Cell 3, 852–862.e4 (2018).
Pastinen, T. Cis-acting regulatory variation in the human genome. Science 306, 647–650 (2004).
Ge, B. et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nature Genet. 41, 1216–1222 (2009).
Pastinen, T. et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics 16, 184–193 (2004).
Morley, M. et al. Genetic analysis of genome-wide variation in human gene expression. Nature 430, 743–747 (2004).
Calabrese, C. et al. Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020).
Rhee, J.-K., Lee, S., Park, W.-Y., Kim, Y.-H. & Kim, T.-M. Allelic imbalance of somatic mutations in cancer genomes and transcriptomes. Sci. Rep. 7, 1653 (2017).
Meyer, K. B. et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 6, e108 (2008).
Maia, A.-T. et al. Extent of differential allelic expression of candidate breast cancer genes is similar in blood and breast. Breast Cancer Res. 11, R88 (2009).
Cox, D. G. et al. Common variants of the BRCA1 wild-type allele modify the risk of breast cancer in BRCA1 mutation carriers. Human Mol. Genet. 20, 4732–4747 (2011).
Maia, A.-T. et al. Effects of BRCA2 cis-regulation in normal breast and cancer risk amongst BRCA2 mutation carriers. Breast Cancer Res. 14, R63 (2012).
Liu, R. et al. Allele-specific expression analysis methods for high-density SNP microarray data. Bioinformatics 28, 1102–1108 (2012).
Xiao, R. & Scott, L. J. Detection of cis-acting regulatory SNPs using allelic expression data. Genetic Epidemiol. 35, 515–525 (2011).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Hartman, D. J., Davison, J. M., Foxwell, T. J., Nikiforova, M. N. & Chiosea, S. I. Mutant allele-specific imbalance modulates prognostic impact of KRAS mutations in colorectal adenocarcinoma and is associated with worse overall survival. Int. J. Cancer 131, 1810–1817 (2012).
Soh, J. et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE 4, e7464 (2009).
Krasinskas, A. M., Moser, A. J., Saka, B., Adsay, N. V. & Chiosea, S. I. KRAS mutant allele-specific imbalance is associated with worse prognosis in pancreatic cancer and progression to undifferentiated carcinoma of the pancreas. Modern Pathol. 26, 1346–1354 (2013).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416.e11 (2018).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Dunnwald, L. K., Rossing, M. A. & Li, C. I. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Research 9, R6 (2007).
Chia, S. et al. Human epidermal growth factor receptor 2 overexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J. Clin. Oncol. 26, 5697–5704 (2008).
Keegan, N. M., Gleeson, J. P., Hennessy, B. T. & Morris, P. G. PI3K inhibition to overcome endocrine resistance in breast cancer. Expert Opin. Investig. Drugs 27, 1–15 (2018).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Yan, H. Allelic variation in human gene expression. Science 297, 1143–1143 (2002).
Huang, F. W. et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013).
Mansour, M. R. et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014).
Przytycki, P. F. & Singh, M. Differential allele-specific expression uncovers breast cancer genes dysregulated by cis noncoding mutations. Cell Syst. 10, 193–203.e4 (2020).
Shoemaker, R., Deng, J., Wang, W. & Zhang, K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 20, 883–889 (2010).
Ongen, H. et al. Putative cis-regulatory drivers in colorectal cancer. Nature 512, 87–90 (2014).
Morgese, F. et al. Impact of phosphoinositide-3-kinase and vitamin D3 nuclear receptor single-nucleotide polymorphisms on the outcome of malignant melanoma patients. Oncotarget 8, 75914–75923 (2017).
Li, Q. et al. Associations between single-nucleotide polymorphisms in the PI3K-PTEN-AKT-mTOR pathway and increased risk of brain metastasis in patients with non-small cell lung cancer. Clin. Cancer Res. 19, 6252–6260 (2013).
Wang, L.-E. et al. Roles of genetic variants in the PI3K and RAS/RAF pathways in susceptibility to endometrial cancer and clinical outcomes. J. Cancer Res. Clin. Oncol.138, 377–385 (2011).
Pu, X. et al. PI3K/PTEN/AKT/mTOR pathway genetic variation predicts toxicity and distant progression in lung cancer patients receiving platinum-based chemotherapy. Lung Cancer 71, 82–88 (2011).
Vasan, N. et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 366, 714–723 (2019).
Wilkerson, M. D. et al. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic Acids Res. 42, e107–e107 (2014).
Verlaan, D. J. et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 19, 118–127 (2009).
Arnold, M., Raffler, J., Pfeufer, A., Suhre, K. & Kastenmüller, G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336 (2014).
Li, Y., Willer, C. J., Ding, J., Scheet, P. & Abecasis, G. R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiol. 34, 816–834 (2010).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2011).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Coetzee, S. G., Coetzee, G. A. & Hazelett, D. J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites: Fig. 1. Bioinformatics btv470 https://doi.org/10.1093/bioinformatics/btv470 (2015).
Xu, H. et al. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1823, 818–825 (2012).
Lees, E., Faha, B., Dulic, V., Reed, S. I. & Harlow, E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 6, 1874–1885 (1992).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2012).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
McLaren, W. et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Heap, G. A. et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Human Mol. Genet. 19, 122–134 (2009).
Castel, S. E., Levy-Moonshine, A., Mohammadi, P., Banks, E. & Lappalainen, T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 16, 195 (2015).
Chen, J. et al. A uniform survey of allele-specific binding and expression over 1000-Genomes-Project individuals. Nat. Commun. 7, 11101 (2016).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signaling 6, pl1 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Andrews, S. FastQC: A QualityControl Tool for High Throughput Sequence Data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Carvalho, B. S., Louis, T. A. & Irizarry, R. A. Quantifying uncertainty in genotype calls. Bioinformatics 26, 242–249 (2009).
Gonzalez, J. R. et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics 23, 654–655 (2007).
Lawrence, M., Gentleman, R. & Carey, V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009).
Deelen, P. et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes 7, 901 (2014).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Human Genet. 81, 559–575 (2007).
Therneau, T. M. & Grambsch, P. M. Modeling Survival Data: Extending the Cox Model (Springer New York, 2000).
Therneau, T. M. A Package for Survival Analysis in R. https://cran.r-project.org/web/packages/survival/ (2021).
Acknowledgements
We thank all the patients who donated tissue and the associated pseudo-anonymized clinical data for this project. The authors would also like to thank the Functional Genomics of Cancer group members at CINTESIS-UAlg for helpful discussions and Vitor Morais at UAIC for administrative support. This work was supported by Portuguese national funding through FCT-Fundação para a Ciência e a Tecnologia, and CRESC ALGARVE 2020, institutional support ALG-01-0145-FEDER-31477—DevoCancer, ALG-01-0145-FEDER-30895—Intergen, CBMR—UID/BIM/04773/2013, CINTESIS R&D Unit—UIDB/4255/2020, POCI-01-0145-FEDER-022184 - GenomePT, the contract DL 57/2016/CP1361/CT0042 (J.M.X.) and individual fellowships SFRH/BPD/99502/2014 (J.M.X.) and PD/BD/114252/2016 (F.E.). Funding was also received from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/303745 (A.T.M.), and a Maratona da Saúde Award (A.T.M.). The METABRIC project was funded by Cancer Research UK, the British Columbia Cancer Foundation, and the Canadian Breast Cancer Foundation BC/Yukon. This sequencing project was funded by CRUK grant C507/A16278 and CRUK core grant A16942. The authors also acknowledge the support of the University of Cambridge, Hutchinson Whampoa, the NIHR Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, the Centre for Translational Genomics (CTAG) Vancouver, and the BCCA Breast Cancer Outcomes Unit. We thank the Genomics, Histopathology, and Biorepository Core Facilities at the Cancer Research UK Cambridge Institute and the Addenbrooke’s Human Research Tissue Bank (supported by the National Institute for Health Research Cambridge Biomedical Research Centre).
Author information
Authors and Affiliations
Contributions
L.C., R.M., J.M.X., S.F.C., and A.T.M. wrote the manuscript. R.M., J.M.X., S.F.C., and A.T.M. contributed to the overall design of this study. Data were collected by F.E., A.B., L.M., R.B., C.C., S.F.C., and A.T.M. Data were analyzed and interpreted by L.C., R.M., J.M.X., B.P.A., F.E., C.S., I.D., M.E., A.M., I.A.S., J.S., and A.T.M. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Correia, L., Magno, R., Xavier, J.M. et al. Allelic expression imbalance of PIK3CA mutations is frequent in breast cancer and prognostically significant. npj Breast Cancer 8, 71 (2022). https://doi.org/10.1038/s41523-022-00435-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-022-00435-9