Abstract
Late-stage relapse (LSR) in patients with breast cancer (BC) occurs more than five years and up to 10 years after initial treatment and has less than 30% 5-year relative survival rate. Long non-coding RNAs (lncRNAs) play important roles in BC yet have not been studied in LSR BC. Here, we identify 1127 lncRNAs differentially expressed in LSR BC via transcriptome sequencing and analysis of 72 early-stage and 24 LSR BC patient tumors. Decreasing expression of the most up-regulated lncRNA, LINC00355, in BC and MCF7 long-term estrogen deprived cell lines decreases cellular invasion and proliferation. Subsequent mechanistic studies show that LINC00355 binds to MENIN and changes occupancy at the CDKN1B promoter to decrease p27Kip. In summary, this is a key study discovering lncRNAs in LSR BC and LINC00355 association with epigenetic regulation and proliferation in BC.
Similar content being viewed by others
Introduction
Breast cancer is the most common cancer diagnosed among US women with ~276,480 estimated new cancer cases in 2020 and is the second leading cause of cancer deaths among women1. The 5-year relative survival for localized disease is above 98%. However, this decreases significantly to 28% for distant metastasis2. Breast cancer relapse occurs within the first 3–5 years after initial treatment; however, those that relapse after five years are termed late-stage relapse (LSR) breast cancer3. Trials of patients treated with adjuvant endocrine therapy for five years have shown a 50% reduction in the risk of relapsing, but the risk of relapse is 10% and as high as 41% depending on initial tumor node status and tumor grade4,5,6. Additionally, there are limited treatment options for patients with LSR breast cancer and few of these patients cannot undergo chemotherapy owing to organ dysfunction or lower performance status as a result of widespread metastasis7. Overall, more studies are needed to understand the benefits of long-term hormone therapy and the underlying molecular and genetic mechanisms promoting LSR.
Long non-coding RNAs (lncRNAs) are greater than 200 nucleotides in length, do not encode proteins8, and have a diverse range of epigenetic and biological functions, including serving in many functions associated with carcinogenesis and metastasis9,10,11,12,13,14. LncRNAs have been found to be deregulated in breast cancer15,16,17,18,19,20,21,22 and have been associated with drug resistance23,24,25. Since lncRNAs serve as potential biomarkers due to their strong tissue specificity26,27, we hypothesize that they can also be used as biomarkers for LSR. Subsequent mechanistic studies could improve our understanding of why some patients relapse more than five years after treatment. However, despite multiple studies identifying the roles of lncRNAs in the distant metastasis of breast cancer23,28,29,30,31, the contribution of lncRNAs to LSR breast cancer has not been explored. This is primarily due to limited availability of LSR patient cohorts for molecular characterization. To overcome this barrier, we utilized ER + early-stage II and III tumor samples accrued from two neoadjuvant aromatase inhibitor treatment trials32,33 and LSR patient tumor samples from Washington University34. Using transcriptome sequencing, we compared the LSR samples to early-stage samples to identify deregulated lncRNAs associated with relapse. We identified LINC00355 to be the most up-regulated lncRNA in LSR breast cancer patient samples and cancer cell lines. Next, we determined that LINC00355 promoted cellular proliferation by binding to the MENIN protein to decrease the expression of cyclin-dependent kinase inhibitor, p27Kip. This study provides the landscape of lncRNAs in LSR and mechanistic evidence of LINC00355 contribution in LSR breast cancer and proliferation.
Results
Identification of long non-coding RNAs in late-stage relapse breast cancer
In order to identify which lncRNAs are associated with LSR, we analyzed transcriptome sequencing data from 72 early-stage patient samples from two preoperative neoadjuvant aromatase inhibitor treatment trials (ACOSOG-Z1031/NCT00265759 and NCT00084396)32,33, termed “early-stage” and 24 LSR patient samples sequenced at Washington University34, termed “late-stage” (Supp. Table 1). Our analysis revealed 1,127 differentially expressed (DE) lncRNAs (FDR < 0.001, absolute log2FC > 2, Supp. Table 2) between the early-stage and late-stage relapse breast cancer patient samples (Fig. 1a, Supp. Fig. 1).
a Schematic of the pipeline used to identify lncRNAs driving late-stage relapse breast cancer. b Heatmap showing the mean expression of patient samples and cell lines for the lncRNAs in late-stage relapse breast cancer. Side bar represent the log fold-change. c Top upregulated gene sets found to be correlated with late-stage relapse as determined by GSEA. d Top GSEA enrichment up-regulated gene sets.
Additionally, due to the complex subtypes in breast cancer, we also evaluated the expression of the DE lncRNAs associated with LSR breast cancer in a panel of publicly available RNA sequence data from 31 nonmalignant and malignant ER + breast cancer cell lines35. We were able to identify 68 deregulated lncRNAs associated with LSR and highly expressed in cell lines with Fragments Per Kilobase of transcript per Million mapped reads (FPKM) > 5 in at least two breast cancer cell lines (Fig. 1a, b, and Supp. Fig. 2). We identified several known lncRNAs previously identified to promote breast cancer including LINC0258236, CASC937,38,39, PDXDC2P40, NNT-AS141, AP000439.342, and lncRNAs found to be expressed in other cancer types (Fig. 1b). Next, we performed a gene set enrichment analysis (GSEA) to evaluate the association of the identified lncRNA genes with cancer gene signatures (Fig. 1c). Our analysis found enrichment of multiple gene sets that were associated with breast cancer and estrogen receptor including MORF_ESR1 (nominal p value = 0.001) and GSEA sets indicating roles in FIRESTEIN_PROLIFERATION (nominal p value = 0.001), and TAVAZOIE_METASTASIS (nominal p value = 0.001, Fig. 1c and d). Using the unbiased approach of transcriptome sequencing and unique patient samples from the two clinical trials of preoperative neoadjuvant aromatase inhibitor treated and late-stage relapse patients, we identified 68 lncRNAs that we believe are associated with late-stage relapse breast cancer signaling and phenotypes.
LINC00355 is the most up-regulated lncRNA in LSR breast cancer
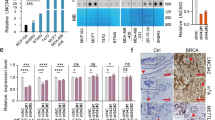
We focused on characterizing the top upregulated lncRNA LINC00355 (NR_145420.1, Fold change= 7.21, p = 2.7e-13), in LSR breast cancer when comparing early-stage patient samples (Figs. 1b and 2a). LINC00355 is a previously annotated 1878 nucleotide long lncRNA first identified as an oncogene in bladder cancer43. Similar to our early-stage patient cohort, LINC00355 was downregulated in 480 early-stage breast cancer samples from The Cancer Genome Atlas (TCGA) from multiple breast cancer subtypes (Fig. 2a): triple negative (n = 72), mean FPKM = 0.151, HER2 + (n = 29), mean FPKM = 0.215, luminal A (n = 298), mean FPKM = 0.237, and luminal B (n = 81), mean FPKM = 0.299, and normal tissue (n = 77), mean FPKM = 0.174. LINC00355 was only highly expressed in the late-stage relapse breast cancer patient samples (mean FPKM = 28.092, Fig. 2a). Additionally, we detected increased LINC00355 expression in publicly available RNA-Seq data35 of malignant breast cell lines compared to non-malignant cell lines (p = 0.047, Fig. 2b, Supp. Fig. 2). To better assess LINC00355 cell type-specific expression and show expression levels are not due to contamination of using bulk patient tissue from RNA-Seq, we also assessed its expression in single-cell RNA-Seq data from breast tissues downloaded from Wu et al.44 We detected very low, 0.90% or less, of cells expressing LINC00355, which was restricted to cancer epithelial cells (Supp. Fig. 3a–c). In contrast, XIST lncRNA is expressed in 35% of cancer epithelial cells and is over 50% expressed in myeloid, cancer-associated fibroblast, and endothelial cells (Supp. Fig. 3a, d, and e). We further assessed expression of LINC00355 in normal tissue using RNA-Seq data from Genotype-Tissue Expression (GTEx) and see slightly higher expression in whole blood, omentum tissue, subcutaneous adipose tissue, and higher expression in EBV-transformed lymphocytes and testis compared to breast mammary tissue (Supp. Fig. 4). Overall, LINC00355 is more highly expressed in LSR compared to early-stage breast cancer patient samples, and in malignant compared to non-malignant cell lines and normal tissues.
a Expression of LINC00355 in primary tumors from the The Cancer Genome Atlas (TCGA) and early-stage and late-stage relapse from patient cohorts. b Sequence expression of LINC00355 in cell line panels and c qPCR from in-house cell lines showing subtypes. Estrogen Receptor (ER), Progesterone receptor (PR), Human epidermal growth factor receptor 2 (HER2), Tumor protein 53 (TP53), Mutation (M) d Nuclear localization of LINC00355 in T47D. MTNR1 (cytoplasmic positive control), U1, and MALAT1 (nuclear positive controls). All data are presented as mean values ± s.d., analyzed by two-tailed paired t-test, and repeated more than two times. Source data are provided as a Source Data File.
Next, we assessed LINC00355 expression in a panel of breast cancer cell lines by quantitative PCR (qPCR) that included both luminal A and luminal B subtype cell lines. LINC00355 is expressed greater than tenfold in breast cancer cell lines compared to the non-tumorigenic cell line MCF10-A (Fig. 2c). Since subcellular localization may provide insight into lncRNA putative functions, we fractionated estrogen receptor positive (ERα) T47D cells, which had highest endogenous levels of LINC00355, and found that LINC00355 is pre-dominantly expressed in the nucleus (Fig. 2d). In summary, we show that LINC00355 previously found to have oncogenic potential is also highly expressed in breast cancer patient samples and cell lines. Specifically, we were able to determine LINC00355 is selectively highly expressed in LSR breast cancer.
LINC00355 expression promotes proliferation and invasion in malignant breast cancer cell lines
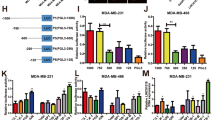
In order to gain a better understanding of the role LINC00355 plays in LSR breast cancer, we transiently silenced its expression with two siRNAs (siRNA1 and siRNA2, Supp. Table 3) in malignant breast cancer cell lines (T47D and CAMA-1) which have high endogenous LINC00355 expression (Fig. 3a). As LINC00355 was previously found to be associated with cellular proliferation45,46,47, we conducted EdU (5-ethynyl-2′-deoxyuridine) proliferation assays using flow cytometry in T47D and CAMA-1 cells with at least 50% silenced LINC00355 expression. We observed a significant decrease of proliferation in T47D cells lines with silenced LINC00355 compared to negative control scrambled siRNAs (siRNA1 p = 4.99e-05, siRNA2 p = 0.006, two-tailed paired t-test; Fig. 3b). Next, we detected a significant decrease in cell viability for 3 days post 72-h LINC00355 knockdown compared to control siRNAs by Alamar Blue Assay (Day 5, siRNA1 p = 0.0007, siRNA2 p = 0.008; Day 6, siRNA1 p = 0.02, siRNA2 p = 0.02, two-tailed paired t test; Supp. Fig. 5a). In addition, we assessed DNA content in T47D cells with siRNAs targeting LINC00355 and detected a decrease in the S phase of cell cycle (siRNA1 p = 0.002, siRNA2 p = 0.0003, two-tailed paired t test, Fig. 3c). A significant decrease in proliferation (siRNA1 p = 0.0002, siRNA2 p = 0.001, two-tailed paired t test, Fig. 3b) and S phase of cell cycle (siRNA1 p = 0.007, siRNA2 p = 0.03, two-tailed paired t test, Fig. 3c) was similarly seen in the second breast cancer cell line CAMA-1.
a Transient knockdown of LINC00355 in T47D and CAMA-1 cell lines. b Decreased LINC00355 expression decreased proliferation measured by EdU incorporation and c decrease in S phase. Decreased LINC00355 expression increased d p27Kip protein and e CDKN1B mRNA levels. Fold change normalized to control. f–h Knockdown expression of LINC00355 decreased cellular invasion. i MCF10A cell line with LINC00355 overexpression and empty vector. j LINC00355 overexpression increased proliferation measured by EdU incorporation and k increase in S phase. LINC00355 overexpression decreased l p27Kip protein and m CDKN1B mRNA levels. Fold change normalized to empty vector. n and o LINC00355 overexpression increased cellular invasion. *p value < 0.05, **p value < 0.005, #p value < 0.0005, no difference (n.d.). All data are presented as mean values ± s.d, analyzed by two-tailed paired t test, and repeated more than two times. Bar = 25 µM, Dapi stained nuclei are shown in white. Source data are provided as a Source Data File.
Due to the importance of the cyclin-dependent kinase inhibitor p27Kip as one of the key regulators of progression from G1 to S phase in cell cycle and its frequent decreased concentration in human malignancies48, we confirmed protein expression of p27Kip in T47D and CAMA-1 cell lines. Silencing LINC00355 increased protein levels of p27Kip in both cell lines (T47D, siRNA1 fold = 1.45, siRNA2 fold 1.79; CAMA-1 siRNA1 fold = 1.97, siRNA2 fold = 2.49, Fig. 3d). There was also more than 1.5-fold increase of CDKN1B mRNA, which encodes transcription for p27Kip in LINC00355 silenced cell lines (Fig. 3e).
Next, we evaluated whether LINC00355 promotes cellular invasion by seeding Matrigel-coated transwells in a modified Boyden chamber assay. We found decreased cellular invasion when LINC00355 is silenced in T47D cells (siRNA1 p = 2.98e-06, siRNA2 p = 4.56e-05, two-tailed paired t test, Fig. 3f and g) and CAMA-1 cells (siRNA1 p = 0.01, siRNA2 p = 0.006, two-tailed paired t test, Fig. 3f, h) compared to negative controls.
Further, we developed a MCF10A stable cell line with more than 11,500-fold LINC00355 overexpression compared to empty vector cells that had negligible LINC00355 expression (Fig. 3i). We similarly detect an increase in proliferation in overexpression cells compared to empty vector by EdU assay (p = 0.002, two-tailed paired t-test, Fig. 3j), Alamar Blue assay (Day 5, p = 0.0006; Day 6, p = 0.00007, two-tailed paired t test; Supp. Fig. 5b), and S phase in cell cycle by assessing DNA content (p = 2.33 e-05, two-tailed paired t test, Fig. 3k). There was also more than 1.5-fold decrease of p27Kip protein levels (Fig. 3l) and CDKN1B mRNA (p = 0.0019, two-tailed paired t test, Fig. 3m) in LINC00355 overexpressed cell lines. Finally, we found increased cellular invasion when LINC00355 is overexpressed when compared to empty vector (p = 0.003, two-tailed paired t test, Fig. 3n and o) by modified Boyden chamber assay. These data indicate that LINC00355 induces proliferation and invasion in cell lines possibly through the regulation of p27Kip.
LINC00355 expression promotes proliferation and invasion in long-term estrogen deprived cell lines
To further assess the metastatic behavior associated LSR breast cancer, we determined if LINC00355 also induces aggressive phenotypes in a late-stage relapse setting by using two ERα + cell lines T47D and MCF7 that were deprived of estrogen for longer than 3 years, termed long-term estrogen deprived (LTED) cells49. The LTED model has been developed to recapitulate the acquired resistance to aromatase inhibitors. However, the two LTED models have independent mechanisms of resistance. The T47D LTED model lacks ESR1 locus amplification whereas the MCF7 LTED model has ESR1 amplification leading to increased ERα protein expression49,50,51. Wild type T47D cell lines when deprived of estrogen, termed T47D LTED, show a loss of ERα protein (Fold = 0.21) compared to its wild type counterpart (Fig. 4a). In contrast, MCF7 cell lines when deprived of estrogen, termed MCF7 LTED, show an amplified ERα protein level (Fold = 1.88) compared to its wild-type counterpart (Fig. 4a). LINC00355 expression was decreased by 65-fold in T47D LTED when compared to wild type T47D cells as measured by qPCR, and conversely there was a 60-fold increase of LINC00355 expression in and MCF7 LTED cell lines compared to wild type MCF7 (Fig. 4b). These results demonstrate an association of LINC00355 with ERα.
a Expression of Estrogen Receptor in T47D and MCF7 wild-type and long-term estrogen deprived (LTED) cell lines. Fold change normalized to respective wild-type cell lines. b LINC00355 expression in primary and LTED cell lines. c Transient knockdown of LINC00355 in MCF7 LTED cell lines. d and e Decreased LINC00355 expression in MCF7 LTED cells decreased proliferation by EdU and S phase. f Decreased LINC00355 expression increased p27Kip protein and g CDKN1B mRNA expression. Fold change normalized to control. h and i Cellular invasion decreased with LINC00355 knockdown. j LINC00355 expression inT47D LTED overexpression cell lines. k and l LINC00355 overexpression in T47D LTED cells increased proliferation by EdU and S phase. m LINC00355 overexpression decreased CDKN1B mRNA expression. n and o Cellular invasion increased with LINC00355 overexpression. *p value < 0.05, **p value < 0.005, #p value < 0.0005, no difference (N.D). Bar = 25 µM. All data are presented as mean values ± s.d, analyzed by two-tailed paired t test, and repeated more than two times. Dapi stained nuclei are shown in white. Source data are provided as a Source Data File.
Due to the increased levels of LINC00355 expression in MCF7 LTED cells, we assessed whether LINC00355 promoted aggressiveness of MCF7 LTED cells. Greater than 70% silencing of LINC00355 in MCF7 LTED cell lines (Fig. 4c) resulted in a significant decrease of cellular proliferation (siRNA1 p = 0.0003, siRNA2 p = 0.02, two-tailed paired t test, Fig. 4d), Alamar Blue assay (Day 5, siRNA1 p = 0.015, siRNA2 p = 0.038; Day 6, siRNA1 p = 0.03, siRNA2 p = 0.0008, two-tailed paired t test; Supp. Fig. 5c), and S phase of cell cycle (siRNA1 p = 0.0006, siRNA2 p = 0.013, two-tailed paired t test, Fig. 4e) in MCF7 LTED cells compared to the negative control scrambled siRNAs. Additionally, we saw more than twofold increase in p27Kip protein levels (Fig. 4f) and more than 1.5-fold increase in CDKN1B mRNA expression (Fig. 4g) in the MCF7 LTED cell line with silenced LINC00355. Further, we evaluated cellular invasion in MCF7 LTED cells lacking expression of LINC00355. We observed a significant decrease of invasion (siRNA1 p = 0.002, siRNA2 p = 0.004, two-tailed paired t test) in MCF7 LTED cells with siRNAs targeting LINC00355 as compared to cells treated with negative control scrambled siRNA (Fig. 4h and i). Last, silenced LINC00355 expression in MCF7 LTED cells then re-introduction of LINC00355 expression, restores invasiveness of MCF7 LTED cells compared to silenced cells (siRNA p = 0.018, siRNA plus overexpression p = 0.012, Supp. Fig. 6).
We overexpressed LINC00355 in the T47D LTED cell lines more than 15.5-fold compared to empty vector cells that had negligible LINC00355 expression (Fig. 4j). We detect an increase in proliferation in T47D LTED overexpressed cells compared to empty vector by EdU assay (p = 0.019, two-tailed paired t test, Fig. 4k) and S phase in cell cycle by assessing DNA content (p = 0.007, two-tailed paired t test, Fig. 4l). There was also more than 60% decrease of CDKN1B mRNA (Fig. 4m) in LINC00355 T47D LTED overexpressed cell lines. We lastly found an increase in cellular invasion in T47D LTED overexpressed cells when compared to empty vector (p = 0.0003, two-tailed paired t test, Fig. 4n and o). Taken together, LINC00355 induces cellular proliferation and invasion in both malignant breast cancer cell lines and long-term estrogen-deprived cell lines that mimic late-stage relapse.
LINC00355 binds to MENIN to regulate CDKN1B expression
Since LINC00355 is localized in the nucleus, increases cellular proliferation, and decreases p27Kip protein levels, we hypothesize that LINC00355 may transcriptionally regulate CDKN1B, the gene that encodes for p27Kip protein. Previously it was shown that MENIN, encoded by MEN1 (multiple endocrine neoplasia 1), is required for its transcriptional activation of p27Kip by increasing histone H3 lysine 4 methylation (H3K4me3) at the promoter of CDKN1B52,53,54,55. Thus, we assessed if LINC00355 may directly bind to MENIN. We first assessed MENIN and ERα protein levels to show that protein levels did not change upon silencing of LINC00355 in both the highly expressed primary cell line T47D and the LSR model MCF7 LTED cell line (Fig. 5a and b). We next conducted an RNA immunoprecipitation coupled with qPCR (RIP-qPCR) with MENIN in the T47D cell line. Indeed, we detected a more than 21-fold enrichment of LINC00355 by RIP-qPCR compared to IgG control and do not see enrichment of XIST RNA, as a negative control of MENIN binding (Fig. 5c). We also show LINC00355 increased MENIN binding in MCF7 LTED cells (Fold change = 10.4 in MENIN compared to IgG; Fig. 5d). To orthogonally validate these findings, we conducted an RNA pull-down assay utilizing a 5′ Bromo-UTP full-length LINC00355 sense labeled probe and a negative control antisense probe to pull-down proteins that may be bound to LINC00355. We found that the LINC00355 sense probe was bound to MENIN protein compared with the control probe (Fig. 5e) by Western blot of nuclear lysates. To identify the regions of LINC00355 that bind to MENIN, we conducted cross-linking immunoprecipitation and qPCR in MCF10A cells transfected with an empty vector and LINC00355 full length (overexpression) using eight primers tiling LINC00355 (Supp. Fig. 7a). We detected Primer 4 and Primer 5 tiling 627-1023 nucleotides had higher fold enrichment (Fold >3 and Fold >5, respectively) of binding to MENIN (Supp. Fig. 7b). We re-validated our previous findings that full-length LINC00355 binds to MENIN around 789 nucleotides (Fig. 5c and d). These orthogonal methods support LINC00355 binding to MENIN protein. To determine the effects of LINC00355-MENIN binding at the promoter of CDKN1B to regulate the expression of p27Kip protein, we silenced LINC00355 to show a significant increase in MENIN occupancy at the CDKN1B promoter in T47D cells (siRNA1 p = 7.33 e -07, siRNA2 p = 0.0006, two-tailed paired t test, Fig. 5f) as compared to IgG control. We also observed an increase in H3K4me3 at the promoter of CDKN1B with silenced LINC00355 (siRNA1 p = 0.004, siRNA2 p = 0.01, two-tailed paired t-test, Fig. 5g). The increase in MENIN and H3K4me3 occupancy was further detected in MCF7 LTED cells with silenced LINC00355 in chromatin immunoprecipitation (ChIP) with MENIN (siRNA1 p = 0.03, siRNA2 p = 0.001) and ChIP with H3K4me3 (siRNA1 p = 0.0001, siRNA2 p = 0.002, two-tailed paired t test, Fig. 5h and i). Decreased MENIN and H3K4me3 occupancy was detected in MCF10A cells with overexpressed LINC00355 (MENIN; p = 0.001, H3K4me3; p = 0.01, two-tailed paired t test, Fig. 5j and k). We additionally identified two other genes regulated by MENIN including AGR356 and FOXA156, that are also shown to be regulated by LINC00355. We show a significant decrease of expression of CDKN1B (p = 0.04), AGR3 (p = 0.005) and FOXA1 (p = 0.02) using a siRNA targeting MEN1 (Supp. Fig. 8). We again validate a significant increase of CDKN1B expression (p = 0.02) with silenced LINC00355 expression and additionally detected an increase of AGR3 (p = 0.02) and FOXA1 (p = 0.004) genes (Supp. Fig. 8). Taken together, we provide evidence that LINC00355 functions by binding to the MENIN protein, which decreases its occupancy at the promoter of CDKN1B, decreasing protein levels of p27Kip, increasing proliferation, and cellular invasion in late-stage relapse breast cancer models (Fig. 6).
Protein expression in a T47D and b MCF7 LTED cells upon LINC00355 knockdown. Estrogen Receptor α (ERα). Fold change normalized to control. LINC00355 RNA immunoprecipitation (RIP) of MENIN in c T47D and d MCF7 LTED cells. e MCF7 LTED RNA pull-down of LINC00355 with MENIN. T47D cell line chromatin immunoprecipitation (ChIP) of f MENIN and g H3K4me3 at the CDKN1B promoter with knock down of LINC00355. MCF7 LTED cell line ChIP of h MENIN and i H3K4me3 at the CDKN1B promoter with knock down of LINC00355. MCF10A overexpression cell line ChIP of j MENIN and k H3K4me3 at the CDKN1B promoter. *p value < 0.05, **p value < 0.005, #p value < 0.0005. All data are presented as mean values ± s.d, analyzed by two-tailed paired t test, and repeated more than two times. Source data are provided as a Source Data File.
MENIN binding to LINC00355 changes its occupancy at the CDKN1B promoter to decrease CDKN1B gene expression and p27Kip protein levels. This alters S phase cell cycle checkpoint that leads to increased cellular proliferation and contributing to phenotypes of late-stage relapse breast cancer. Created with Biorender.com.
Discussion
Although most relapses occur during the first 5 years after breast cancer diagnosis about 30% of ER + positive breast cancer patients relapse more than 5 years and up to 10 years after their 5-year endocrine therapy treatment. Since distant metastases are responsible for the majority of breast cancer deaths, finding ways to reduce the risk of distant metastases associated with late relapse is critical in improving survival rates from the disease. Our study used ER + early-stage (II and III) tumor tissues isolated from two neoadjuvant aromatase inhibitor (AI) therapy trials. This unique set of tumor samples allowed us to assess lncRNA expression in a preoperative AI treatment setting that is currently being assessed as a new treatment option. Additionally, we incorporated a unique very limited cohort of LSR patients. Thus, our study provides a systematic transcriptome analysis of lncRNAs expression in early-stage and late-stage relapse breast cancer patients to detect deregulated lncRNAs significantly altered in LSR breast cancer.
Through our analysis of lncRNAs in LSR breast cancer, we prioritized the most up-regulated lncRNA, LINC00355. Notably, LINC00355 was previously reported to promote proliferation in multiple cancer types including bladder cancer, colorectal cancer, prostate cancer, lung adenocarcinoma, and head and neck squamous cell carcinoma43,45,46,47,57,58,59 and induce epithelial to mesenchymal transcription, and serve as a diagnostic biomarker in colon cancer45,46,47,57,58,59,60; however, it has not yet been identified in breast cancer and identified here specifically in ER + late-stage relapse breast cancer. High nuclear enrichment of LINC00355 was also previously detected in bladder cancer43, which we confirmed in breast cancer. This led us to hypothesize that LINC00355 may also play a role in inducing proliferation in LSR breast cancer through transcriptional regulation of key genes. Interestingly, we found that LINC00355 induced proliferation and cellular invasion in malignant breast cancer cell lines. We also utilized MCF7 and T47D long-term estrogen receptor deprived cells lines that mimic late-stage relapse patient samples as they have been deprived of estrogen for longer than three years and have previously been shown to provide an in vitro parallel of patients treated with aromatase inhibitor having post-treatment loss or amplification of estrogen receptor61. When using the LTED cell lines, we showed LINC00355 induced proliferation and cellular invasion. Because the MCF7 LTED cell line has amplified estrogen receptor protein, we have also provided evidence that LINC00355 may have a role in an ER + amplified setting, a hypothesis that needs further investigation.
As LINC00355 was able to induce proliferation, we decided to investigate a key cell signaling regulator, p27Kip. p27Kip signaling is highly complex and has multiple modes of both transcriptional and post-transcriptional regulation62. Indeed, we found that our breast cancer cell lines with high levels of LINC00355 also had decreased expression p27Kip. Moreover, we determined that LINC00355 expression is also inversely correlated with CDKN1B gene expression. This indicated that LINC00355 might transcriptionally regulate CDKN1B to alter cellular proliferation. We hypothesized that LINC00355 may interact with a negative regulator of cell cycle separating the protein from its native target, thus causing an uncontrolled increase in cellular proliferation. There are numerous well-known examples of lncRNAs that function as negative regulators or decoys including: CISAL63, ROR64, PANDA65, and MEG366. Previous research determined that the CDKN1B gene is transcriptionally negatively regulated by MENIN in ER + breast cancer56 through epigenetic modifications and maintenance of transcription at multiple loci for cell cycle regulators52,54,67. Given this, we focused our study on MENIN to determine if LINC00355 interacts with MENIN sequestering it from the promoter of CDKN1B gene leading to decreased p27Kip protein levels and disruption of the cell cycle control and increasing proliferation. This study indicates a lncRNA to bind to MENIN which provides evidence of the important functional regulation of lncRNAs. An important unanswered question is the stoichiometry of LINC00355 in regulating the association of MENIN with the CDKN1B locus. Further, we highlight the importance of lncRNA expression in LSR breast cancer that may be used as novel therapies in the future. In conclusion, our study provides a landscape of lncRNAs in LSR and provides key evidence of their contribution to LSR breast cancer.
Methods
Clinical samples and cell lines
The primary breast cancer samples for this study were accrued from two neoadjuvant endocrine therapy trials32,33. RNA-Seq data accrued from two neoadjuvant aromatase inhibitor therapy trials (ACOSOG-Z1031, NCT00084396, and NCT00265759)32,33,34 are available via the dbGAP database (with accession phs000472). The studies were supported by the Clinical Trials Support Unit and approved by the institutional review boards of all participating institutions; all patients provided signed informed consent. Twenty-four late-stage relapse patients were enrolled and consented under a banking protocol approved by the Washington University School of Medicine Institutional Review Board (approval number 201102244, Supp. Table 1), their tumors were resected for RNA sequencing. PolyA RNA was isolated and created into Illumina TruSeq libraries run on Illumina HiSeq 2000 platform (accession GSE189389). The RNA-Seq gene expression data (FPKM) and the clinical data of TCGA BRCA samples were downloaded from the TCGA data portal. RNA-Seq data of the 32 breast cancer cell lines were obtained from the NCBI Gene Expression Omnibus (accession GSE48216)35. All cell lines with “unknown” subtype were removed and a representative number of cell lines from each subtype were retained.
Sequence alignment and transcript quantification
All sequencing reads from both patient and cell lines data were aligned to the human reference genome hg19 Ensembl release 75 using TopHat version v2.0.868. TopHat was run according to an in-house pipeline at The McDonnell Genome Institute (bowtie-version = 2.1.0, library-type fr-unstranded, mate-inner-dis 254, and mate-std-dev 50). The remaining TopHat parameters were left to their default values. For accurate alignment, transcriptome index file (transcript sequences) was provided to guide the alignment. Raw read counts for transcripts were generated using featureCounts version v1.4.6-p369 and were used to compute transcript expression levels as normalized in FPKM (Fragments Per Kilobase of transcript per Million mapped reads) format.
RNA-Sequencing data analysis
Differential expression analysis was performed between early-stage and late-stage relapse samples using the negative binomial generalized log-linear model and likelihood ratio tests capabilities of edgeR version v3.8.670 using the raw read counts. To correct for batch effect, RUVSeq R package version v1.0.071 was used with a list of negative control genes72. To enhance our confidence in the differentially expressed genes we discovered, lowly expressed transcripts were removed and only transcripts with expression > 1 FPKM in at least 50% of samples in either group were retained for downstream analysis. All transcripts with FDR < 0.001 and absolute log fold change > 2 were considered differentially expressed transcripts. TCGA subtypes and cell lines expression difference significance was calculated specifically for LINC00355 using the nonparametric Wilcoxon rank-sum test.
Gene enrichment analysis
To identify gene signatures that are associated with the identified differentially expressed genes, a gene set enrichment analysis (GSEA)73 was performed on the list of up and down regulated lncRNAs with 1000 gene set permutations using Signal2Noise metric. Significantly enriched gene sets were determined by false discovery rate (FDR) adjusted P ≤0.1 and normalized enrichment score (NES) ≥ 1.5.
Cell Culture
All breast cancer cell lines were a kind gift from Dr. Matthew Ellis and Jieya Shao at Washington University in St. Louis. T47D, MCF7, HCC1428, BT483, ZR75B, and HCC1500 cell lines were grown in RPMI 1640 media (Invitrogen, Carlsbad, CA), 10% fetal bovine serum (Sigma, St. Louis, MO), 1% HEPES (Sigma), 0.5% glucose (Sigma), 1% sodium pyruvate (Sigma), 1% L-glutamine (Sigma), and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). T47D LTED and MCF7 LTED cells were grown in phenol red free RPMI 1640 media (Invitrogen), 10% Charcoal stripped bovine serum (Sigma), 1% HEPES, 0.5% glucose, 1% sodium pyruvate, 1% L-glutamine, and 1% penicillin/streptomycin. CAMA-1 and BT-474 cells were grown in DMEM (Invitrogen), 10% fetal bovine serum, and 1% penicillin/streptomycin. MDA175 were grown in DMEM F12 (Invitrogen), 10% fetal bovin serum, and 1% penicillin/streptomycin. MCF10A cells were grown in DMEM F12 (Invitrogen), 10% horse serum serum, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 20 ng/mL EGF and 1% penicillin/streptomycin. We silenced expression of LINC00355 using custom silencer select RNAs (siRNAs) targeting LINC00355 or Silencer Select Negative Control No.1 siRNA (Thermofisher, Austin, TX). siRNA sequences are listed in Supplementary Table 3.
Full-length LINC00355 transcript was PCR amplified from T47D cells and cloned into the pCFG5-IEGZ vector (a kind gift from Dr. Ron Bose, Washington University). Full-length LINC00355 inserts were confirmed with Sanger sequencing at GeneWiz. Retroviral infection of cells was performed according to Kauri et al.74. Briefly, the amyotrophic phoenix cell line was transfected with 10 μg of pCFG5-LINC00355 or empty vector control by calcium phosphate precipitation and incubated for 24-h. Viral supernatants were harvested after an additional 24-h incubation. Virus was added to cells seeded in six-well dishes in the presence of 8 μg/mL polybrene (Sigma), centrifuged at 300 Å~ g for 90 min, and fresh media was added to the plate. After 14 days of Zeocin (Invitrogen) selection cells were used for assays. MCF10A cells or T47D LTED cells that had low endogenous expression of LINC00355 were infected with virus expressing LINC00355 or empty vector for 48 h and selected with 100 μg/mL Zeocin.
Nuclear cytoplasmic isolations
Nuclear and cytoplasmic isolations were conducted using the PARIS Kit (Thermo Fisher, Waltham, MO) following the manufacturer’s protocol. Total RNA was collected as described below. Nuclear and cytoplasmic isolations were calculated by normalizing respective gene to total RNA expression.
Quantitative real-time PCR
Total RNA was isolated for each breast cancer cell line using NucleoSpin RNA plus with DNA removal column (Macherey-Nagel, Duren). Total RNA was then transcribed to cDNA with SuperScript III First-strand cDNA system (Invitrogen) to verify expression of genes and verify knockdown efficiencies using Fast SyberGreen Master Mix (Invitrogen) as per the manufacturer’s protocol. Primer sequences are available in Supplementary Table 3.
Modified Boyden chamber assay
Cell lines were seeded at 350,000 cells in a six-well dish. The next day cells were transfected at 50 nM with two independent custom designed siRNAs or a negative scramble control (Supplementary Table 3) with Lipofectamine RNAiMax (Invitrogen) for 72-h or 2 µg pCFG5-LINC00355 or empty vector control with Lipofectamine 3000 (Invitrogen) for 72-h. Cells were then harvested and re-seeded in complete media at 200,000 cells on an 8.0 µM permeable membrane support transwell (Corning, Corning, NY) pre-coated with 200 µg/mL Matrigel (Corning) in 24-well plates creating a modified Boyden chamber assay. A serum gradient was established with cells plated in serum-free media added to the bottom of the well. Cells were allowed to invade overnight and then fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA). Next, nuclei were stained with DAPI (Sigma) (1 µg/µL). A cotton swab was used to remove non-invading cells from the top of the membrane. Invaded DAPI-stained cells were then imaged with Q-Capture Pro software on an Olympus IX70 microscope, quantified using ImageJ software (http://imagej.nih.gov/ij/), and statistical significance was determined by a student’s t test. Five or more images were taken per transwell membrane at 20× magnification. Assays were repeated two to three times.
Proliferation assay
Cell lines were seeded at 350,000 cells in a six-well dish. The next day cells were transfected at 50 nM with siRNAs targeting LINC00355 or negative control pCFG5-LINC00355 or empty vector control as described above in Modified Boyden chamber assay section. Seventy-two hours later cells were pulsed with EdU (5-ethynyl-2′-deoxyuridine) (Thermo Fisher, Carlsbad, CA, cat# C10420) for 3 h and harvested by trypsinization. Cells were then fixed, permeabilized, and washed following manufacturer’s instructions. Cells were stained for DNA content with FxCycle Violet (Thermo). Analysis of EdU and cell cycle was assessed by measuring DNA content on a flow cytometer machine (FACScan, Becton Dickinson) at the Siteman Cancer Center Flow Cytometry Core. We collected a minimum of 25,000 cells per sample in triplicate. FlowJo Version10 (Becton Dickinson) was used to analyze data.
Western blot
Protein was collected by plating 300,00–350,000 representative cancer cells in a six-well dish. Cells were transfected as described above. Cells were then lysed with Tris Lysis buffer (50 mM TrisHCl, 1% Triton X-100, 131 mM NaCl, 1 mM sodium orthovanadate, 10 mM Na4P207, 10 mM NaF, 1 mM EDTA, and proteasome inhibitor), run on an agarose gel and transferred to nitrocellulose membranes. Blots were then probed overnight at four degrees with respective antibodies. All antibodies and concentrations are listed in Supplementary Table 4. Blots were then washed with TBST buffer and then applied with secondary goat anti-rabbit HRP linked or goat anti-mouse HRP-linked antibodies (Thermo Fisher, Waltham, MA). Lastly, blots were washed, visualized with Clarity Western ECL Substrate (BioRad, Hercules, CA) and imaged using the ChemiDoc XRS + System (BioRad). Blots were derived from the same experiment. Raw western blots are shown in Supplementary Fig. 9 were processed in parallel and derived from the same experiment.
RNA immunoprecipitation (RIP)
RIP coupled to qPCR assays were conducted by isolating nuclear lysates from ten million T47D or MCF7 LTED cells following the NER-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Fisher). Nuclear lysates were then incubated overnight rotating with 5 µg of Anti-Menin antibody or IgG antibody isotype control in RIPA wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1% NP40, 0.5% Na-Deoxycholate, 0.05% SDS, 1 mM EDTA) and SUPERase-in RNAse inhibitor (Invitrogen). The next day 50 µL of Invitrogen Dynabeads Protein G were added to the antibody lysate/mixture and rotated for 1–2 h at 4 °C. Next, beads were washed six times with RIPA wash buffer using a magnetic bead separator. Protein was then digested with Proteinase K buffer (RIPA buffer, 10% SDS, 10 mg/ml Proteinase K), at 55 °C for 30 min shaking. RNA was phenol:chloroform:isoamyl alcohol extracted following the general protocol (Thermo Fisher). Last, gDNA was removed from RNA using ArticZymes Heat and Run gDNA removal kit following the manufacturer’s protocol (Tromso, Norway). cDNA was made using SuperScript III First strand cDNA system as indicated above and qPCR was run with Fast SyberGreen MasterMix and indicated primers (Supplementary Table 3). Fold enrichment of qPCR results were calculated following Sigma-Aldrich Data Analysis Calculation Shell by comparing non-specific control IgG antibody raw CTs to MENIN or H3K4me3 RNA binding protein CT normalized against 1% input.
BrU-labeled RNA pull-down
Full-length LINC00355 RNA probes were made using the Promega Riboprobe in vitro transcription kit from 2.5 μg of linearized DNA in the pGEM-3Z vector (Madison, WI). Control antisense probes were made by in vitro transcription from the SP6 promoter. LINC00355 RNA pull-down experiments were performed in MCF7 LTED nuclear lysates following the RiboTrap Kit manufacturer’s protocol (MBL, Woburn, MA).
Chromatin immunoprecipitation (ChIP)
ChIP coupled to qPCR assays were conducted by first sonicating five million cells in SDS lysis buffer (1% SDS, 500 mM EDTA, 50 mM Tris-HCl pH8). Next, immunoprecipitation with 5 µg of IgG, MENIN, or H3K4me3 antibodies was done by incubating sonicated lysate with indicated antibody in ChIP Dilution Buffer (0.01% SDS, 1.10% Triton X-100, 1.2 nM EDTA, 16.7 mM Tris-HCl pH8, 167 mM NaCl), and 1X Halt Protease and Phosphatase inhibitors overnight with rotation at 4°. The next day 50ul of Dynabeads Protein G (Invitrogen) were added to the antibody lysate mixture and rotated for 1 h. Bead/lysate mixture was then washed once with Low Salt Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH8, 150 mM NaCl), then High Salt Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH8, 500 mM NaCl), Lithium Chloride Wash Buffer (0.25 M lithium chloride, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH8), and finally two washes with Tris-HCl EDTA Buffer (10 mM Tris-HCl pH8, 1 mM EDTA). DNA was eluted by incubating beads for 30 min at room temperature with SDS Elution Buffer (1% SDS, 0.1 M sodium bicarbonate) followed by 1.25 M NaCl and 2.5 mg/ml RNAse A at 95° for 15 min shaking followed by addition of Proteinase K buffer (1 µL 10 mg/ml Proteinase K, 5 µM 0.5uL EDTA, 10 µL 1 M Tris pH7.5) shaking at 60° for 15 min. DNA was then isolated using phenol:chloroform:isoamyl alcohol extraction following the general protocol as mentioned above. DNA was diluted by five and used for qPCR. The % input calculation was determined by comparing CT values from input DNA and ChIP DNA for the CDKN1B target promoter region using the following equation: %Input = % of starting input fraction × 2^[CT(input)−CT(ChIP)]. Primer sequences are available in Supplementary Table 3.
Single-cell RNA sequencing analysis
Breast single-cell data from 26 patients using the 10x chromium platform was downloaded from Gene Expression Omnibus (GSE176078) from Wu et al.44 Seurat v4.1.0 (Butler et al.75; Hafemeister and Satija76) was used for all subsequent analyses. We applied a series of quality filters to the data to remove barcodes which fell into any one of the following categories recommended by Seurat: transcript counts below 300; total genes expressed below 200 and above 10,000; UMI count below 1000 or above 10,000; mitochondrial gene expression larger than 10%. The Seurat object was constructed using the published gene and feature matrices across the sample cohort. The dataset was scaled and normalized and corrected for batch effects using Seurat’s “SCTransform” function (regressing by nCount_RNA and percent of mitochondrial DNA, variable.features n = 2000). Cells were clustered using the Louvain algorithm (Blondel et al.,77) and top 30 PCA dimensions using the following functions: “FindNeighbors” and “FindClusters” (resolution = 0.5). The resulting merged and normalized matrix was used for the subsequent analysis. Cells were annotated using the published metadata from the original publication.
Alamar blue assay
Cell lines were seeded at 350,000 cells in a six-well dish. The next day cells were transfected at 50 nM with two independent custom designed siRNAs or a negative scramble control (Supplementary Table 3) with Lipofectamine RNAiMax (Invitrogen) for 72 h or 2 µg pCFG5-LINC00355 overexpression or empty vector control with Lipofectamine 3000 (Invitrogen) for 72 h. Cells were then harvested and re-seeded in complete media in 96-well plates of cells. The plates were incubated for 3 days. Percent viability was scored by incubating cells for 3-h with AlamarBlue HS Cell Viability reagent (Invitrogen A50100). The reaction was stopped by the addition of 1% SDS. Fluorescence Ex/Em 540/590 was read in a Varioscan Lux plate reader. The fluorescence values for the vehicle plates were averaged and percent viability was determined by the formula: Percent viability = (average vehicle − value)/(average vehicle − average resazurin in media blank) Å~ 100.
Cross-linking immunoprecipitation (CLIP)
Prospective cells are seeded at twenty million in 150 cm dish. The next day cells are washed with 15 ml ice cold PBS twice and adjusted to 10mls per dish. Dishes are uncovered and irradiated with 150 mJ/cm2 of UVA (254 nm) in Stratalinker. Cells are then harvested and centrifuged at 2000 RPM at 4 °C for 5 min. Cell pellets are resuspended by adding 1 ml of NP-40 lysis buffer (20 mM Tris–HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40) with 1ul protease inhibitors, and 1 mM DTT then incubated on ice for 10 min and centrifuge at 10,000 RPM for 15 min at 4 °C. Supernatants were collected and 1U/μl RNase T1 was added then incubated at 22 C for 30 min. 35ul of 5 M EDTA was added to stop reaction. Protein G Beads were washed two times with ice-cold PBS per sample and resuspended in 100uls NT2 buffer (50 mM Tris–HCl at pH 7.5, 150 mM NaCl,1 mM MgCl2, 0.05% NP-40) with 5ug of respective antibody, then subsequent rotation for 1 h at room temperature. All antibodies and concentrations are listed in Supplementary Table 4. Beads are then washed with NT2 buffer to remove excess antibody. Lysates are then added to beads for three hours at 4 °C, washed and incubated with 20 units RNAse-free DNase I for 15 min at 37 °C thermomixer shaking slowly. Protein kinase buffer (141 uls NP-40 lysis buffer, 0.1% SDS, 0.5 mg/ml Proteinase K) is then added and incubated for 15 min at 55 °C on thermomixer shaking at max speed. Supernate is then collected and isolation of RNA is conducted using standard phenol:cholorform:isoamyl alchohol protocol.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The early-stage breast cancer RNA sequencing data referenced in this study are available from dbGaP database under the accession code phs000472 and the late-stage relapse breast cancer relapse RNA-Seq data generated in this study are available in the NCBI Gene Expression Omnibus (GEO) under the accession code GSE189389. The RNA-Seq data of the breast cancer cell lines referenced in this the study are available in a public repository from the NCBI GEO under the accession code GSE48213. The data used for the GTEx analyses described in this manuscript were obtained from the GTEx Portal (accession number phs000424.vN.pN) on 11/18/21. The source data underlying figures are provided as a Source Data file. All the other data supporting the findings of this study are available within the article, its supplementary information files, and from the corresponding author upon request. A reporting summary for this article is available as a Supplementary Information file.
References
Siegel, R. L. et al. Colorectal cancer statistics, 2020. CA: Cancer J. Clin. 70, 145–164 (2020).
Cronin, K. A. et al. Annual report to the nation on the status of cancer, part I: National Cancer Statistics. Cancer 124, 2785–2800 (2018).
Omidvari, S. et al. Very late relapse in breast cancer survivors: a report of 6 cases. Iran. J. Cancer Prev. 6, 113–117 (2013).
Pan, H. et al. Predictors of recurrence during years 5-14 in 46,138 women with ER+ breast cancer allocated 5 years only of endocrine therapy (ET). J. Clin. Oncol. 34, 505–505 (2016).
Pan, H. et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 377, 1836–1846 (2017).
Early Breast Cancer Trialists’ Collaborative, G. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 (2011).
Wangchinda, P. & Ithimakin, S. Factors that predict recurrence later than 5 years after initial treatment in operable breast cancer. World J. Surg. Oncol. 14, 223 (2016).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Weidle, U. H., Birzele, F., Kollmorgen, G. & Ruger, R. Long non-coding RNAs and their role in metastasis. Cancer Genom. Proteom. 14, 143–160 (2017).
Gooding, A. J. et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci. Rep. 7, 12698 (2017).
Silva, J. & Smith, D. Long Non-Coding RNAs and Cancer (Caister Academic Press, 2012).
Silva-Fisher, J. M. et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat. Commun. 11, 2156 (2020).
Dang, H. X. et al. Long non-coding RNA LCAL62 / LINC00261 is associated with lung adenocarcinoma prognosis. Heliyon 6, e03521 (2020).
White, N. M. et al. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur. Urol. 71, 257–266 (2017).
Zhang, T. et al. Long non-coding RNA and breast cancer. Technol. Cancer Res. Treat. 18, 1533033819843889 (2019).
Jia, Y. et al. Transcriptome sequencing profiles reveal lncRNAs may involve in breast cancer (ER/PR positive type) by interaction with RAS-associated genes. Pathol. Res. Pract. 215, 152405 (2019).
Shao, X. et al. MicroRNA and LncRNA expression profiles in human estrogen receptor-positive breast cancer. Clin. Lab. 65, https://doi.org/10.7754/Clin.Lab.2018.180340 (2019).
Mathias, C., Zambalde, E. P., Rask, P., Gradia, D. F. & de Oliveira, J. C. Long non-coding RNAs differential expression in breast cancer subtypes: what do we know? Clin. Genet. 95, 558–568 (2019).
Silva, J. M., Boczek, N. J., Berres, M. W., Ma, X. & Smith, D. I. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 8, 496–505 (2011).
Silva, J. M. et al. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 95, 355–362 (2010).
Arun, G. et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016).
Han, Y. J. et al. LncRNA BLAT1 is upregulated in basal-like breast cancer through epigenetic modifications. Sci. Rep. 8, 15572 (2018).
Wu, Y. et al. The role of lncRNAs in the distant metastasis of breast cancer. Front. Oncol. 9, 407 (2019).
Yousefi, H. et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 39, 953–974 (2020).
Zhou, S. et al. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci. Rep. 38, https://doi.org/10.1042/BSR20180772 (2018).
Fan, C. N., Ma, L. & Liu, N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J. Transl. Med. 16, 264 (2018).
Vishnubalaji, R., Shaath, H., Elkord, E. & Alajez, N. M. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFbeta and focal adhesion kinase (FAK) signaling. Cell Death Discov. 5, 109 (2019).
Hou, L. et al. Long noncoding RNA ROR promotes breast cancer by regulating the TGF-beta pathway. Cancer Cell Int. 18, 142 (2018).
Xiu, B. et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 18, 187 (2019).
Kim, J. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 (2018).
Tan, B. S. et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38, 5612–5626 (2019).
Ellis, M. J. et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype-ACOSOG Z1031. J. Clin. Oncol. 29, 2342–Z2349 (2011).
Olson, J. A. Jr et al. Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: results from a multicenter phase II trial. J. Am. Coll. Surg. 208, 906–914 (2009). discussion 915-906.
Lei, J. T. et al. Functional annotation of ESR1 gene fusions in estrogen receptor-positive breast cancer. Cell Rep. 24, 1434–1444 e1437 (2018).
Daemen, A. et al. Modeling precision treatment of breast cancer. Genome Biol. 14, R110 (2013).
Wang, B. et al. Long noncoding RNA LINC02582 acts downstream of miR-200c to promote radioresistance through CHK1 in breast cancer cells. Cell Death Dis. 10, 764 (2019).
Jiang, B. et al. Long noncoding RNA cancer susceptibility candidate 9 promotes doxorubicinresistant breast cancer by binding to enhancer of zeste homolog 2. Int. J. Mol. Med. 42, 2801–2810 (2018).
Zhang, J., Wang, Q. & Quan, Z. Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-beta expression. Biochem. Biophys. Res. Commun. 515, 644–650 (2019).
Shao, G. et al. lncRNA CASC9 positively regulates CHK1 to promote breast cancer cell proliferation and survival through sponging the miR195/497 cluster. Int. J. Oncol. 54, 1665–1675 (2019).
Katayama, M. L. H. et al. Stromal cell signature associated with response to neoadjuvant chemotherapy in locally advanced breast cancer. Cells 8, https://doi.org/10.3390/cells8121566 (2019).
Li, Y. et al. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed. Pharmacother. 103, 939–946 (2018).
Zhang, Y. et al. Genome-wide study of ER-regulated lncRNAs shows AP000439.3 may function as a key regulator of cell cycle in breast cancer. Oncol. Rep. 38, 3227–3237 (2017).
Seitz, A. K. et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci. Rep. 7, 395 (2017).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021).
Liang, Y. et al. A novel long non-coding RNA LINC00355 promotes proliferation of lung adenocarcinoma cells by down-regulating miR-195 and up-regulating the expression of CCNE1. Cell. Signal. 66, 109462 (2020).
Yan, L., Wang, P., Fang, W. & Liang, C. Cancer-associated fibroblasts-derived exosomes-mediated transfer of LINC00355 regulates bladder cancer cell proliferation and invasion. Cell Biochem. Funct., https://doi.org/10.1002/cbf.3462 (2019).
Lu, S., Sun, Z., Tang, L. & Chen, L. LINC00355 promotes tumor progression in HNSCC by hindering MicroRNA-195-mediated suppression of HOXA10 expression. Mol. Ther. Nucleic acids 19, 61–71 (2020).
Alkarain, A. & Slingerland, J. Deregulation of p27 by oncogenic signaling and its prognostic significance in breast cancer. Breast Cancer Res. 6, 13–21 (2004).
Li, S. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
Aguilar, H. et al. Biological reprogramming in acquired resistance to endocrine therapy of breast cancer. Oncogene 29, 6071–6083 (2010).
Masri, S. et al. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 68, 4910–4918 (2008).
Alrezk, R., Hannah-Shmouni, F. & Stratakis, C. A. MEN4 and CDKN1B mutations: the latest of the MEN syndromes. Endocr.-Relat. Cancer 24, T195–T208 (2017).
Pellegata, N. S. et al. Germ-line mutations in p27Kip1 cause a multiple endocrine neoplasia syndrome in rats and humans. Proc. Natl Acad. Sci. USA 103, 15558–15563 (2006).
Karnik, S. K. et al. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc. Natl Acad. Sci. USA 102, 14659–14664 (2005).
Milne, T. A. et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl Acad. Sci. USA 102, 749–754 (2005).
Dreijerink, K. M. A. et al. Enhancer-mediated oncogenic function of the Menin tumor suppressor in breast cancer. Cell Rep. 18, 2359–2372 (2017).
Jiang, T. et al. Identification of potential prostate cancer-related pseudogenes based on competitive endogenous RNA network hypothesis. Med. Sci. Monit. 24, 4213–4239 (2018).
Yuan, W. et al. Comprehensive analysis of lncRNA-associated ceRNA network in colorectal cancer. Biochem. Biophys. Res. Commun. 508, 374–379 (2019).
Zhang, Z. et al. Analysis of lncRNA-associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell. Physiol. Biochem. 49, 1778–1791 (2018).
Yazarlou, F. et al. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag. Res. 10, 6357–6365 (2018).
Sanchez, C. G. et al. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 13, R21 (2011).
Hnit, S. S. et al. p27(Kip1) signaling: transcriptional and post-translational regulation. Int. J. Biochem. Cell Biol. 68, 9–14 (2015).
Fan, S. et al. lncRNA CISAL inhibits BRCA1 transcription by forming a tertiary structure at its promoter. iScience 23, 100835 (2020).
Fan, J. et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 16, 139 (2015).
Peng, C. et al. Over expression of long non-coding RNA PANDA promotes hepatocellular carcinoma by inhibiting senescence associated inflammatory factor IL8. Sci. Rep. 7, 4186 (2017).
Li, R. et al. MEG3-4 is a miRNA decoy that regulates IL-1beta abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci. Signal. 11, https://doi.org/10.1126/scisignal.aao2387 (2018).
Hughes, C. M. et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13, 587–597 (2004).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
Eisenberg, E. & Levanon, E. Y. Human housekeeping genes, revisited. Trends Genet. 29, 569–574 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kavuri, S. M. et al. Cellular FLICE-inhibitory protein (cFLIP) isoforms block CD95- and TRAIL death receptor-induced gene induction irrespective of processing of caspase-8 or cFLIP in the death-inducing signaling complex. J. Biol. Chem. 286, 16631–16646 (2011).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Blondel, V., Guillaume, J., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 83, 10008 (2008).
Acknowledgements
J.S.F. received funding from the Washington University School of Medicine Molecular Oncology Training Grant (T32CA113275) and Faculty Diversity Scholar Award. C.A.M. received funding from the Susan G. Komen Career Catalyst Award. M.J.E. received funding from a Susan G. Komen Promise Grant (PG12220321), a Cancer Prevention Institute of Texas (CPRIT) Recruitment of Established Investigators Award (RR140033), the AVON Foundation and the McNair Foundation, the Breast Cancer Research Foundation (BCRF ELFF-16-003), and R01-CA095614. J.S.R-F. is funded in part by the by a Cancer Center Support Grant of the National Institutes of Health (NIH)/ National Cancer Institute (grant No P30CA008748), by an NIH/NCI P50 CA247749 01 grant and by a Breast Cancer Research Foundation grant. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO., for the use of the Siteman Flow Cytometry that provided flow cytometry service. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Author information
Authors and Affiliations
Contributions
J.S.F. and C.A.M. designed and directed experimental studies and are co-senior authors. A.M.E., A.M., J.T.L, R.G.J., and H.X.D., performed sequencing data analysis. J.S.F., P.K.T., K.Z.G., C.Y.T., E.B.R., and N.M.W. performed experimental studies. M.J.E. provided samples. M.J.E., C.A.M., and J.S.F. provided financial support. J.S.R.-F., E.R.M., and L.D. provided project guidance. J.S.F., A.M.E., and C.A.M. wrote manuscript, which all authors reviewed.
Corresponding authors
Ethics declarations
Competing interests
J.S.R-F. reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics and Paige.AI, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics, Paige.AI and Personalis, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro, outside the scope of this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eteleeb, A.M., Thunuguntla, P.K., Gelev, K.Z. et al. LINC00355 regulates p27KIP expression by binding to MENIN to induce proliferation in late-stage relapse breast cancer. npj Breast Cancer 8, 49 (2022). https://doi.org/10.1038/s41523-022-00412-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-022-00412-2
This article is cited by
-
Roles of non-coding RNAs in the metabolism and pathogenesis of bladder cancer
Human Cell (2023)
-
Apoptosis evasion via long non-coding RNAs in colorectal cancer
Cancer Cell International (2022)