Abstract
In rod-shaped bacteria, morphological plasticity occurs in response to stress, which blocks cell division to promote filamentation. We demonstrate here that overexpression of the patatin-like phospholipase variant CapVQ329R, but not CapV, causes pronounced sulA-independent pyridoxine-inhibited cell filamentation in the Escherichia coli K-12-derivative MG1655 associated with restriction of flagella production and swimming motility. Conserved amino acids in canonical patatin-like phospholipase A motifs, but not the nucleophilic serine, are required to mediate CapVQ329R phenotypes. Furthermore, CapVQ329R production substantially alters the lipidome and colony morphotype including rdar biofilm formation with modulation of the production of the biofilm activator CsgD, and affects additional bacterial traits such as the efficiency of phage infection and antimicrobial susceptibility. Moreover, genetically diverse commensal and pathogenic E. coli strains and Salmonella typhimurium responded with cell filamentation and modulation in colony morphotype formation to CapVQ329R expression. In conclusion, this work identifies the CapV variant CapVQ329R as a pleiotropic regulator, emphasizes a scaffold function for patatin-like phospholipases, and highlights the impact of the substitution of a single conserved amino acid for protein functionality and alteration of host physiology.
Similar content being viewed by others
Introduction
Bacteria are defined by their cell shape such as rod, coccus, spiral or filamentous, which exist as unicellular or multinucleated cells1. Shaped differently, many bacterial species display extensive morphological plasticity in response to environmental cues including severe stress2,3. Such morphological variation is often reversible, suggesting an altered physiological cell status or epigenetic modulation of the genetic information upon signal perception or manifested threat. For example, to escape from intracellular biofilms during late infection of bladder cells, rod-shaped uropathogenic Escherichia coli (UPEC) can reversibly transform from a rod with a length of 2-4 μm into a filament of up to 70 μm in length4. In nature, Caulobacter crescentus, a freshwater curved rod-shaped bacterium, can develop 20 μm long helical filaments which escape from biofilms for nutrient acquisition5,6. Other types of filamentous bacteria of diverse phylogenetic origin are commonly found in activated sludge and as host-dependent immunomodulatory ‘segmented filamentous bacteria’ in the gut7,8. The giant Epulopiscium fishelsoni can become up to 750 μm long9.
Upon exposure to various environmental stresses including nutrient deficiency6,10, antibiotic treatment11,12,13, and DNA damage14,15,16, bacterial cell filamentation is a consequence of activation of the SOS response system. This is accomplished mainly by impairment of the functionality of the essential cell division protein, the tubulin homolog FtsZ, which initiates cell division by forming a septal ring at the prospective invagination site upon polymerization17,18,19. Negative regulation of septum forming FtsZ can be exerted by expression of the SOS factor SulA which interacts with FtsZ to inhibit polymerization eventually leading to cell filamentation18,20,21,22. The subsequent arrest in growth enables the cells to recover from otherwise deleterious damages before resuming growth. However, there exist alternative SOS responses and sulA-independent stress-associated pathways that promote filamentation in E. coli. For example, upon treatment with cationic antimicrobial peptides, QueE, an enzyme involved in the queuosine tRNA modification pathway, blocks cell division and induces filamentation23.
Temporal emergence of filamentation can serve alternative purposes such as being a survival strategy contributing to pathogenesis. E. coli filamentation triggered by treatment with cell wall inhibiting β-lactam antibiotics in patients can promote bacterial surface colonization24,25. Filamentation also slows down phagocytosis of UPEC E. coli by macrophages during infection to enhance bacterial survival upon challenge by the host immune responses4,20,26.
During cell division, cell elongation, chromosome replication and segregation, and cytokinesis (cell separation) are coordinated27,28,29 though dysregulation of critical components and regulators promotes cell filamentation and filament formation28,30,31,32,33. For example, overexpression of the cell division inhibitor MinC, which prevents FtsZ septum formation at the cell poles by oscillation from pole to pole assisted by MinDE leads to filamentation34. DamX, a membrane-spanning protein with a peptidoglycan binding SPOR domain, is required for reversible filamentation, colonization and pathogenesis of UPEC morphotype switching35.
Motility is defined as the ability to actively move in liquid or on surfaces36. Various bacterial modes of movement are dependent on energetic requirements with diverse structural elements involved37. As a common mode of motility, propelling of flagellar filaments moves bacterial cells in liquid medium by swimming motility and on a surface of the semi-solid medium by swarming motility36,38. In E. coli and the gastrointestinal pathogen Salmonella enterica serovar Typhimurium, bacterial motility is tightly regulated by global regulatory signals including the second messenger cyclic di-GMP (c-di-GMP) and protein-protein interactions via enzymatically incompetent EAL domain proteins with catalytically active homologs to be involved in c-di-GMP hydrolysis38,39. Cyclic di-GMP is a bacterial intracellular messenger that ubiquitously modulates the single-cell lifestyle transition between motility and sessility for biofilm formation40,41. While flagella-based swimming and swarming motility are post-translationally inhibited by c-di-GMP targeting flagella motor functionality38, c-di-GMP promotes expression of csgD encoding a major transcriptional biofilm regulator, and subsequently the rdar (red, dry, and rough) colony biofilm morphotype is developed.
Besides the ubiquitous c-di-GMP signaling system, other recently identified cyclic dinucleotide second messengers can modulate sessility/motility lifestyle transition in E. coli. The cyclic dinucleotide cyclase DncV synthesizes 3′3′-cyclic AMP-GMP (cAMP-GMP)42,43 to subsequently repress motility and rdar biofilm formation in the animal commensal strain E. coli ECOR3143. DncV is part of the putative capV-dncV-vc0180-vc0181 operon located adjacently to the Yersinia high-pathogenicity island (HPI)44,45. Thereby, capV codes for a cAMP-GMP activated patatin-like phospholipase to induce growth retardation and cytolysis in V. cholerae46,47. Patatin-like phospholipases, which can be found in organisms throughout the phylogenetic tree, hydrolyze the sn-2 ester bond of phospholipids and neutral lipids for lipid turnover, remodeling of membrane composition, and signaling to induce programmed cell death48. These processes are highly regulated and often involve a co-factor to promote enzymatic activity.
In this study, we show that overexpression of the patatin-like phospholipase variant CapVQ329R causes substantial alteration in cell morphology leading to sulA-independent cell filamentation and restricted swimming motility with premature flagella loss. Besides these single-cell morphological changes, which can occur in commensal and pathogenic strains of E. coli of diverse genetic background and S. typhimurium, CapVQ329R expression altered (rdar) colony biofilm formation and susceptibility to antimicrobials and phage infection. These findings demonstrate that a single amino acid change in a patatin-like phospholipase promotes rapid evolution of protein functionality which leads to the enhanced manipulation of various aspects of bacterial physiology.
Results
CapVQ329R inhibits swimming motility of E. coli MG1655
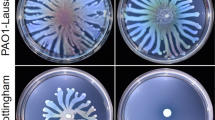
The dinucleotide cyclase DncV synthesizes cAMP-GMP to inhibit rdar biofilm formation and motility in the animal commensal strain E. coli ECOR3149. To assess whether the downstream cAMP-GMP receptor, the patatin-like phospholipase CapV, has a function on its own, we expressed CapV in the heterologous host MG1655, an E. coli K-12 derivative which is not known to synthesize cAMP-GMP nor to harbor DncV. Besides wild-type CapV, we overexpressed its variant CapVQ329R, which had been derived by cloning a respective mutated DNA fragment as described in Supplementary Results. To this end, we observed that expression of the variant CapVQ329R caused suppression of swimming motility, whereas overexpression of the wild-type protein CapV did not alter the apparent motility in semi-solid tryptone broth (TB) agar (as described in Supplementary Results; Fig. 1a and Supplementary Figs. 1–3). TB medium promotes motility of E. coli MG1655 compared to LB medium in the semi-solid agar plate assay50. To further characterize CapVQ329R-induced E. coli MG1655 swimming inhibition, we assessed the production of cell-associated flagella and flagellin upon overexpression of CapVQ329R compared to CapV. Visualization of bacterial cells by transmission electron microscopy (TEM) showed that overexpression of CapVQ329R compared to wild-type CapV dramatically reduced the total number of flagella-producing cells as well as the number of flagella per cell after 6 h incubation at 37 °C (Fig. 1c, d). In agreement, visualization of flagella by Leifson staining upon CapVQ329R overexpression showed cells with intact flagella up to 4 h incubation at 37 °C, but almost no cell with flagella after 6 h (Fig. 1c, e). In congruence with the analysis by TEM and light microscopy examination of Leifson staining, we observed inhibition of production of cell-associated extracellular flagellin in a protein gel upon overexpression of CapVQ329R after 6 h (Fig. 1f, g). Initial analysis of differential gene expression by qRT-PCR indicated 8% downregulation of expression of the class 1 flagella regulon gene flhD encoding a subunit of the major regulator FlhD4C2, and less than 50% downregulation for the representative genes of class 2 fliA encoding the flagella specific sigma factor and class 3 fliC encoding the subunit of flagella, upon CapVQ329R compared to CapV overexpression. Compared to the vector control, flhD, fliA and fliC were downregulated 28, 83, and 73%, respectively, upon overexpression of CapV. Thus, the expression of CapVQ329R interferes with flagella expression beyond the flagella regulon cascade and can affect, for example, depolymerization or degradation of flagella. Cumulatively, these results indicate that CapVQ329R suppressed swimming motility of MG1655 by post-translationally inhibiting the production of flagellar filaments gradually during the growth phase, while initially production of functional flagellar filaments had been observed.
a, b Flagella-dependent swimming motility of wild-type E. coli MG1655 vector control (VC) and upon overexpression of wild-type CapV, its mutants (red) and mutants of CapVQ329R (blue). In total, 3 µl of a OD600 = 5 cell suspension were inoculated into soft agar plates containing 1% tryptone, 0.5% NaCl, and 0.25% agar, and the swimming diameter was measured after 6 h at 37 °C. c Flagella production of a representative E. coli MG1655 VC cell and upon overexpression of CapV, CapVQ329R (Q329R) and CapVQ329R/D197A (D197A) as observed by TEM. d Quantification of the number of flagella per cell upon overexpression of CapV and CapVQ329R after visualization by TEM (c). The number of evaluated cells n = 20. Cells were grown in TB medium for 6 h at 37 °C. e Production of surface-associated flagellin of E. coli MG1655 VC, upon overexpression of CapV and CapVQ329R. f Assessment of flagellin subunit FliC expression by colloidal Coomassie staining from E. coli MG1655 culture supernatants after shearing of flagella upon expression of CapV, CapVQ329R, CapVQ329R/D197A and CapVQ329R/S64A. Cells were grown in TB medium at 37 °C for 6 h. g Assessment of flagellin subunit FliC expression over time in E. coli MG1655 overexpressing CapVQ329R. Samples were harvested at different time points in the growth phase for Western blot analysis of FliC. lc, loading control. h Proposed development of filamentation and flagella inhibition of E. coli MG1655 upon CapVQ329R expression over time. Bars represent mean values from three biologically independent replicates with error bars to represent SD. Differences between mean values were assessed by two-tailed Student’s t test: ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to E. coli MG1655 VC. Vector control VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28. S64A = CapV Q329R/S64A cloned in pBAD28. D197A = CapV Q329R/D197A cloned in pBAD28. R27A = CapVQ329R/R27A cloned in pBAD28. G24A/G25A = CapVQ329R/G24A/G25A cloned in pBAD28.
CapVQ329R expression promotes cell filamentation of E. coli MG1655
Significantly, upon overexpression of CapVQ329R in E. coli MG1655 TEM and light microscopy demonstrated not only the loss of flagella, but the concomitant development of long thin filamentous cells (Fig. 1c, e). In contrast, E. coli MG1655 cells overexpressing wild-type CapV were only slightly elongated compared to the control (Fig. 1c, e).
Assessment of the temporal development of cell filamentation throughout the growth phase upon induced expression of CapVQ329R by light microscopy after Leifson staining indicated that filamentation did not initiate before 2 h after commencement of CapVQ329R expression (Figs. 1e and 2a, b). Subsequently, though the cell length and the frequency of filamentation dramatically increased, whereby after 3 h almost all cells displayed as short filaments around 6 times the length of standard rod cells (Fig. 2a, b). After 4 h of induction, CapVQ329R expressing E. coli MG1655 cells were on average 25 times longer than control cells. After 6 h of induction, CapVQ329R expressing E. coli MG1655 cells were on average more than 50 times longer than the standard rod-shaped cell (Fig. 2, b). Those long cells did not show any movement, while shorter filaments up to approximately 20 times the length of standard E. coli cells, although rare, still showed active swimming motility (Supplementary Movie S1). At the opposite, overexpression of wild-type CapV only slightly increased the cell length compared to E. coli MG1655 control. On note, after 22 h induction of CapVQ329R expression by 0.1% L-arabinose, short rod-shaped motile cells dominated again, which suggested filaments to restart cell division after CapVQ329R expression had diminished due to L-arabinose depletion. In line with this hypothesis, induction of CapVQ329R production by 0.2% l-arabinose did not cause reversion to rod-shaped cells nor showed the cells any movement after 22 h (Fig. 2a, c and Supplementary Movie S2). Upon transfer of those filamentous cells to fresh TB medium without L-arabinose, however, the emergence of rod-shaped cells was again observed (Fig. 2c). A scheme of this developmental process leading to filamentation with consecutive loss of flagella upon expression of CapVQ329R is displayed in Fig. 1h.
a Light microscopy pictures of cell morphology of E. coli MG1655 VC and upon overexpression of CapV and CapVQ329R (Q329R) in TB medium at different time points at 37 °C. b Quantification of cell length of E. coli MG1655 VC and upon overexpression of CapV and CapVQ329R in TB medium at different time points. The quantification is based on results from at least three independent experiments with the assessment of 70 cells from each group. c Cell morphology 3 h after addition of fresh TB medium to filamentous E. coli MG1655 cells overexpressing CapVQ329R. Arrowheads indicate invaginations at proposed future division sites. d, e, f Assessment of cell length upon overexpression of CapV, CapVQ329R and CapVQ329R derivatives in E. coli MG1655 upon induction with different l-arabinose concentrations. Light microscopy pictures (d), quantification of cell length (e) and protein expression level (lc = loading control) (f) upon induction with 0.01% and 0.1% l-arabinose in TB at 37 °C for 4 h. The quantification is based on results from at least three independent experiments with the assessment of 70 cells from each group. Bar, 5 µm. Vector control VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28. G24A/G25A = CapVQ329R/G24A/G25A cloned in pBAD28. R27A = CapVQ329R/R27A cloned in pBAD28. S64A = CapVQ329R/S64A cloned in pBAD28. D197A = CapVQ329R/D197A cloned in pBAD28.
A positive correlation of the filamentation phenotype of the E. coli MG1655 cells with CapVQ329R production level was demonstrated using increasing L-arabinose concentrations. When incubated for 4 h with 0.01% l-arabinose, the low-level CapVQ329R expression created a heterogenous cell population displaying no or restricted cell filamentation (Fig. 2d, e). In contrast upon incubation with 0.1% l-arabinose all cells became filamentous as observed previously. Concomitantly, the CapVQ329R expression level was comparable to the CapV expression level excluding that the observed morphological changes were due to significantly different protein expression levels. As expected, CapV expression was higher upon induction with 0.1% l-arabinose than with 0.01% l-arabinose (Fig. 2f). The 6xHis-tag added to the C-terminus of the protein to detect protein production level did not alter the proficiency of CapVQ329R to induce filamentation (Supplementary Fig. 2g). Cumulatively, filamentation and motility repression are caused by the Q320R substitution in CapV.
CapV is a patatin-like phospholipase, which alters the steady-state lipid profile
Blast search with CapV from E. coli ECOR31 showed that CapV homologs with >60% identity are not only found in individual E. coli and V. cholerae strains, but are widely distributed among gamma-proteobacteria, including Yersinia, Salmonella, Pseudomonas, Shewanella, and Klebsiella species (Supplementary Fig. 4a). Phylogenetic analysis of representative CapV homologs supported classification into four different subgroups (Supplementary Fig. 4b). Alignment of the amino acid sequences of those CapV homologs showed that Q329 is a nearly invariant amino acid even among distantly related CapV proteins (Supplementary Fig. 4a). Q329 is, though, not required for catalytic activity and not part of other characteristic patatin-like phospholipase A2 (PNPLA) consensus motifs and the PNPLA core domain which is restricted to aa 19–210 (Prosite) in the 361 aa long protein. In order to clarify the position of R329 in the protein, we generated a structural model of CapV using the closest structural homolog from the PDB database as a template, the lysophospholipase-like protein FabD from Solanum cardiophyllum (PDB: 1oxwC; Fig. 3a) which shows 22% amino acid identity with CapV. According to this model, R329 is located within helix 12, the second last α helix of CapVQ329R in the context of the RARGRR329 sequence pointing outward with no obvious change in the overall structure of the monomer or potential oligomer assembly to be observed.
a Predicted structural model of the CapV from E. coli ECOR31 shown as ribbon representation. The structural model was built with the I-TASSER server, the result was processed with SWISS-MODEL. The model was based on the coordinates of the 22% identical protein FabD from Solanum cardiophyllum (PDB: 1oxwC). b The graphical representation and schematic indication of the positions of the conserved motifs (indicated by the green bar) and putative active site residues S64 and D197 (marked by red stars) in the PNPLA domain of the 361 aa CapV from E. coli ECOR31 (from L19 to F210). Black arrow, Q329. The graph was assessed by ExPASy_Prosite. c Sequence alignment of CapV from E. coli ECOR31 and selected known phospholipases from other species establishes the conserved motifs of the PNPLA domain, G–G-G-x-[K/R]-G, G-x-S-x-G, and D–G-[A/G], boxed in black, green, and purple, respectively. Entirely conserved residues are shown in white on a red background. Conserved residues are boxed. Putative catalytic residues of CapV are indicated with filled red triangles. The residues in CapVQ329R mutated to alanine are marked with red asterisks above the sequence. The consensus sequence at the bottom indicates in uppercase letter residues with 100% identity and in lowercase letter residues with higher than 70% conservation. Alignment was performed using CLUSTALW, and the result was processed with ESPript 3.0. Sequence identity as in the Methods section. d 32P-cAMP-GMP-DRaCALA of E. coli cell lysates expressing CapV, CapVQ329R, and CapVQ329R/D197A. VC = pBAD28; pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28. D197A = CapVQ329R/D197A cloned in pBAD28. e Assessment of affinity for cAMP-GMP of CapV, CapVQ329R, and CapVQ329R/D197A expressed in E. coli MG1655. 32P-cAMP-GMP was mixed with twofold dilutions of cell extracts starting at a fourfold dilution.
As CapV from V. cholerae46, CapV of E. coli ECOR31 contains a N-terminal canonical PNPLA domain with three main characteristic conserved signature motifs48,51, the phosphate or anion binding motif G–G-G-x-[K/R]-G, the esterase box G-x-S-x-G, and the D–G-[A/G] motif as part of the catalytic dyad (Fig. 3b, c). The G-x-S-x-G motif includes the conserved nucleophilic serine 64 of the active site characteristic for the phospholipase A2 (PLA2) superfamily48,52.
To investigate if catalysis is required for swimming inhibition and filamentation upon CapVQ329R overexpression, a catalytically inactive S64A variant of the protein (CapVQ329R/S64A) was generated. Compared with CapVQ329R, overexpression of CapVQ329R/S64A equally inhibited swimming motility and induced filamentation (Figs. 1a and 2d, e), demonstrating that the G-x-S-x-G motif of CapVQ329R is not required for the phenotype. However, the substitution of arginine in the G–G-G-x-[K/R]-G motif (CapVQ329R/R27A) and aspartic acid of the D–G-[A/G] motif (CapVQ329R/D197A) by alanine relieved both the repression of swimming motility and induction of filamentation. Substitution of the two structural glycine residues (CapVQ329R/G24A/G25A) of the G–G-G-x-[K/R]-G also partially suppressed swimming motility and induced only a mild filamentous phenotype upon overexpression of the protein (Figs. 1a, b and 2d, e). In summary, the G–G-G-x-[K/R]-G and D–G-[A/G] motifs of CapVQ329R are required for repression of the swimming phenotype and cell filamentation.
As substitution of the nucleophilic serine 64 did not relieve motility and cell filamentation, we were wondering whether alternative serine residues are involved in the physiological activity of CapVQ329R. To this end, we substituted S33, S113/114, S146, S177, and S206 by alanine residues. Most of these serine residues were selected as they are located close to the catalytic site (Supplementary Fig. 2f). Only serine 206 was required to induce motility inhibition and promote cell filamentation by CapVQ329R (Supplementary Fig. 2f).
Furthermore, we wanted to clarify whether specifically the Q329R mutation is required to induce filamentation. Replacement of Q329 by the other positively charged amino acid lysine still partially repressed the apparent swimming motility and induced filamentation (Fig. 1b), while replacement of Q329 by asparagine retained the wild-type CapV phenotype (Supplementary Fig. 2f, g).
The binding site for cAMP-GMP in CapV has not been identified. The Q329R substitution might alter the binding of cAMP-GMP or other cyclic dinucleotides to CapV. To this end, binding of cAMP-GMP to CapV and CapVQ329R was analyzed by the differential radial capillary action of ligand (DRaCALA) assay. DRaCALA is based on the retention of small molecular compounds at the protein application spot on a nitrocellulose membrane upon binding. The experiment showed that both proteins bound cAMP-GMP with approximately equal affinity while the CapVQ329R/D197A mutant showed diminished binding (Fig. 3d).
Patatin-like phospholipases hydrolyze the sn-2 acyl ester bond of neutral and phospholipids48,53. In order to assess whether overexpression of CapV and CapVQ329R caused significant changes in the lipid profile concomitant with filamentation, we extracted lipids after 4 h of growth extracts to mass spectrometry (Fig. 4). Based on untargeted charged surface hybrid column quadrupole time-of-flight mass spectrometry (CSH-QTOF MS) analysis a total of 326 lipid species were identified (Fig. 4a). Principle component analysis showed distinct classification of samples into groups correlating with the overexpression of the wild-type CapV and CapVQ329R variant protein (Fig. 4b), indicating that the lipid profiles are significantly altered. Subsequently, we applied hierarchal clustering analysis to segregate the samples cumulatively according to overall changes in the individual lipid compounds. Based on the changes in the lipid compounds, the samples can again be classified into distinct groups according to the expressed proteins (Fig. 4f). Lipids known to be most abundant in the E. coli membrane, phosphatidylethanolamines (PE), phosphatidyl-glycerols (PG) and fatty acids (FAs), displayed the highest relative peak intensity (Fig. 4c–e and Supplementary Fig. 5). We observed significant changes in the peak intensity of members of phospholipid classes, most abundant membrane components of E. coli such as PEs and PGs, but also of free FAs, lysophospholipids (LPE and LPG), phosphatidylcholines (PCs), ceramides (Cer) and sphingomyelins (SM), although the peak intensity of the latter three classes was at least 100-fold lower (Fig. 4c–e and Supplementary Fig. 5). The peak intensity was, however, in none of the cases on the average more than sixfold different. Notably, among the top 50 most significantly altered lipids, PE and PG derivatives with distinct FAs profiles are predominantly represented (Fig. 4f). While CapV overexpression showed downregulation of a restricted group of lipids, a significantly higher number of lipids species were downregulated upon CapVQ329R overexpression. Notably, lipids species were also upregulated upon overexpression of CapV and CapVQ329R suggesting a role of the patatin-like phospholipases in membrane reorganization and/or signaling. Only a few lipid species were distinctively upregulated upon overexpression of CapVQ329R, PE32:2 (16:1 16:1) with two monounsaturated fatty acids and the monounsaturated fatty acid FA18:1. Thus CapV and CapVQ329R might have unique substrate profiles. Whether the observed alterations in the lipid profile are based on distinct residual catalytic activities of the two proteins or indirectly associated with the expression of CapV and CapVQ329R needs to be investigated further. In summary, these results indicate that amino acids in the catalytic motifs are required to induce filamentation and to repress motility. Not or only partially activated CapV and CapVQ329R can alter the lipid profile indicating that CapV is not only a receptor for a second messenger molecule, but might also be involved in alternative second messenger signaling in its nonactivated state.
a The number of identified lipid species in CapV and CapVQ329R induced filamentous cells compared to E. coli MG1655 vector control (VC) by untargeted charged surface hybrid column-quadrupole time-of-flight mass spectrometry (CSH-QTOF MS) analysis. For abbreviation of lipid compounds consult “Methods”. b Principle component analysis of lipid abundance upon overexpression of wild-type CapV and CapVQ329R variant proteins in E. coli MG1655 compared to VC. Of six samples each, outliers have been removed. c–e Alternation and relative abundance of PE (c), PG (d), and FA (e) derivatives by untargeted CSH-QTOF MS analysis. Bars represent mean values from five independent replicates with error bars to represent SD. f Heatmap of selected 50 most significantly altered lipid species built based on hierarchical clustering. Each square represents one sample of each group. The color scale presenting the difference of each log2 transformed peak intensity value to the log2 transformed mean for each lipid species and the percentage of each lipid class is indicated on the right of the heatmap. Heatmap analysis was performed on the Tutools platform (https://www.cloudtutu.com), a free online data analysis website. Differences between mean values were assessed by two-tailed Student’s t test: ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001; black stars in c, d, e and f: compared to MG1655 VC; red stars in f: statistical significance between E. coli MG1655 pCapV and E. coli MG1655 pCapVQ329R. VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28.
CapVQ329R induces asymmetrically positioned FtsZ rings and abnormal nucleoids in filamentous cells
During cell division, positioning of the FtsZ cytokinetic ring at the site of constriction between nucleoids is coordinated with chromosome replication, nucleoid segregation and cell elongation31,54,55. Impairment of this process leads to cell division arrest and filamentation, which can be induced by DNA damage and nucleoid occlusion15,27. After completion of cell segregation, the nucleoid subsequently becomes more compact56. To determine the effect of CapV and CapVQ329R overexpression during the cell division process, we analyzed FtsZ-ring positioning in an E. coli K-12 MG1655 derivative with a chromosomally encoded FtsZ-GFP fusion protein57 and the position and shape of the nucleoid with DAPI staining. After induction of CapV and CapVQ329R in liquid medium at 37 °C for 4 h, we immediately subjected the cells to fluorescence microscopy on agarose pads to visualize cell shape, septum, and FtsZ-ring formation and nucleoid location. Overexpressing wild-type CapV in the E. coli K-12 MG1655 FtsZ-GFP strain, we observed clearly visible constrictions that corresponded with a correctly positioned FtsZ ring and a single nucleoid in nondividing cells and two fully replicated and/or segregated nucleoids in dividing cells, respectively (Fig. 5a). Of note, nucleoids upon CapV expression appeared slightly more compact than those in the control.
a Phase-contrast and fluorescence images of FtsZ-GFP expressing cells (E. coli MG1655 derivative BS001 harboring vector control (VC), pCapV and pCapVQ329R). Cells were cultured in TB medium at 37 °C for 4 h, stained with DAPI, and assessed immediately under fluorescence microscopy. Large fragments of unsegregated nucleoids are indicated by white arrows. Bar, 3 µm. b Quantification of the average distance between two adjacent FtsZ rings upon overexpression of CapV and CapVQ329R, refers to Table 1. c Time-lapse analysis of mCherry-MinC expressing cells (E. coli MG1655 derivative PB318 harboring VC, pCapV and pCapVQ329R). A representative elongated cell is displayed. Graphs on the right of fluorescence images display the line profiles of fluorescent signals emanating from the cell. Arbitrary fluorescent units are obtained, analyzed by the Fiji ImageJ 1.8.0 software, and are plotted on the y axis; cell length (in µm) is plotted on the x axis. Bar, 3 µm. VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28.
In contrast, upon CapVQ329R-induced cell elongation, most filamentous cells displayed smooth contours with no visible septa (Fig. 5a and Supplementary Fig. 6a, b), suggesting that the block in cell division occurs prior to the development of constriction. However, one or two constrictions were occasionally observed in a few filaments (Supplementary Fig. 6a). We found that most of the CapVQ329R-induced filamentous cells were polynuclear and contained multiple abnormally shaped or positioned nucleoids. Discrete patches of DNA staining could be compact, asymmetrically positioned in the filament or displayed an extended or decondensed shape occupying an extensive part of the filament (Fig. 5a and Supplementary Fig. 6b). We also observed a few filamentous cells containing nucleoids, which were evenly positioned throughout the filament with larger interchromosomal spaces (Supplementary Fig. 6b). Collectively, these observations suggest filamentous cells induced by CapVQ329R overexpression to be (partially) defective in chromosome segregation and nucleoid condensation.
Distinct FtsZ rings were observed in most of the filaments suggesting that the block in cell division occurred after FtsZ-ring formation. Large variations in the distance between two FtsZ rings lead to an unequal number of FtsZ rings in filamentous cells of similar length. CapVQ329R promoted filamentation with an average cell length of 31.2 µm (n = 102), which corresponds to >15 times the standard cell length. The majority of the filamentous cells contained three FtsZ rings with multiple segregated and/or unsegregated nucleoids distributed between the rings. Of note, the distance between two adjacent Z-rings varied dramatically in the population of filamentous cells, with on average fourfold longer spacing in CapVQ329R expressing cells compared to cells expressing CapV and the control (Fig. 5b and Table 1). Equally variable was the number of partitioned nucleoids among filamentous cells.
Abnormal FtsZ expression levels induce cell filamentation58. Comparative immunoblot analysis observed slightly lower FtsZ protein production levels in CapVQ329R and CapV overexpressing cells, while the ftsZ mRNA steady-state level was with 91 and 97% of vector control expression only slightly changed (Supplementary Fig. 6c). Whether this (partial) decrease in FtsZ production levels is responsible for CapVQ329R-induced filamentation needs to be further investigated. In summary, CapVQ329R inhibits cell division at the level of constriction initiation and interferes with nucleoid segregation and condensation.
CapVQ329R-induced cell filamentation is sulA-independent
Previous studies showed that SulA blocks cell division by direct interaction with cell division core component FtsZ during the bacterial SOS response19,32. To investigate whether CapVQ329R induced cell filamentation of E. coli MG1655 is caused by the activation of sulA, we analyzed cell morphology upon CapVQ329R overexpression in a sulA mutant59,60. We found that CapVQ329R induced the same filamentous cell phenotype in the sulA mutant as in wild-type E. coli MG1655 (Supplementary Fig. 6d), suggesting that CapVQ329R-induced cell filamentation is sulA-independent consistent with the observed FtsZ-ring formation.
MinC oscillation takes place in filaments
Multiple factors coordinate the regulation of cell division29. Among them is MinC, which oscillates from pole to pole in order to prevent FtsZ-ring formation at the cellular poles34. Upon overexpression, MinC causes cell filamentation. We investigated the dynamics of MinC mobility by time-lapse fluorescence microscopy (Fig. 5c and Supplementary Movie 3). In rod-shaped wild-type cells, we observed that MinC oscillates between cell poles, moving a large fraction of the total fluorescence signals along the long axis of the cell as described previously61. A complete oscillation cycle lasted about 50 s under our experimental conditions. Of note, we found that oscillation of the FtsZ-ring inhibitor MinC is not affected in CapVQ329R-induced filaments (Fig. 5c) (PB318,61). MinC displayed various fluorescence spots ranging from one up to 5 depending on the length of the filament (Fig. 5c and Supplementary Fig. 6e). These fluorescent spots oscillated within multiple invisible cell borders in the filaments suggesting physical restrictions by septa. Within one single filamentous cell, the number of the apparent fluorescence fractions followed thereby a three-step principle: n n − 1 n (n ≧2, located at both poles) or n n + 1 n (n ≧1, located within the cell), during an oscillation periodicity of about 50 s, which is similar as for rod-shaped control cells and wild-type CapV overexpressing cells (Fig. 5c and Supplementary Fig. 6e). It will be relevant to investigate how long-distance MinC oscillation is maintained in filamentous E. coli cells. In summary, MinC oscillation might remain the effective watchdog for the prevention of FtsZ-ring formation in filamentous cells.
Overexpression of CapVQ329R reduces cell numbers
CapV has been identified as a patatin-like phospholipase that causes growth retardation upon activation by cAMP-GMP in V. cholerae El Tor46. In E. coli MG1655, though overexpression of CapV of E. coli ECOR31 wild-type did not affect cell division during the entire growth phase. Though no effect was observed during the first 3 h of cell growth in liquid culture, overexpression of CapVQ329R induced a mild decrease in optical density after 6 h (Fig. 6a). As extensive filamentation might not permit a direct correlation between OD and cell number, we tested cell viability by spotting E. coli MG1655 cells on agar plates for counting of viable individual cells. Compared to the control and cells expressing CapV, ~50% of the colonies were recovered from E. coli MG1655 cell cultures expressing CapVQ329R (Fig. 6b). Consistently, cells stained with the nucleic acid stain SYTO 9 for viability and propidium iodide (PI) for cell death showed that CapVQ329R production induced approximately 50% E. coli MG1655 cells to stain selectively with PI after 6 h, while no DNA staining with PI was observed after 3 h (Fig. 6c and Supplementary Fig. 7a, b). Of note, cells in some filaments took up both dyes seemingly live and dead (Supplementary Fig. 7). Interestingly, the uptake of SYTO 9 into individual cells varied widely in particular in CapV expressing cells, which showed no decrease in cell viability. These data indicate a highly heterogeneous nucleic acid content in individual cells. Thus, expression of not or only partially activated CapV and CapVQ329R might differentially affect membrane permeability and cell viability.
a Growth curves of E. coli MG1655 upon CapV and CapVQ329R overexpression induced by 0.1% l-arabinose in TB at 37 °C. Each data point represents the mean ± SD of six biological replicates. tb = TB medium. b Colony-spotting assay on agar plates. Cells were grown at 37 °C and harvested at different time points. Cell viability determined by spotting serial dilutions (100–10−6) on LB plates to assess colony-forming units. c Quantification of Live/Dead staining of E. coli MG1655 cells after 3 h and 6 h in TB medium at 37 °C. n = 1200. VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28.
Vitamin B6 restricts cell filamentation induced by CapVQ329R
Filamentation has been shown to be affected by environmental conditions26. We found that filamentation was particularly pronounced during cell growth in TB medium (1% tryptone, 0.5% NaCl), while it was restricted when cultured in LB medium (1% tryptone, 1% NaCl, 0.5% yeast extract) (Fig. 7a). Supplementation of TB medium with 0.5% yeast extract (YE) repressed the filamentous phenotype dramatically, while 5% restored the rod shape of all cells. Supplementation with 0.5% NaCl did not affect filamentation.
a Light microscopy pictures of E. coli MG1655 cell morphology and, b quantification of cell length in LB and TB medium supplemented with 0.5% NaCl, 0.5% YE, 5% YE, and VB6 (pyridoxine, 5 mg/ml), respectively. The quantification is based on results from at least three independent experiments with the assessment of 70 cells from each group. Bar, 5 µm. VC = pBAD28. Q329R = CapVQ329R cloned in pBAD28.
Yeast extract is the water-soluble portion of autolyzed yeast cells used to prepare microbiological culture media62. As a nutrient source, it provides nitrogen, amino acids, peptides, carbohydrates, vitamin B complex, and other components that promote microbial growth63. In particular, yeast extract contains B vitamins, water-soluble precursors of structurally unrelated enzyme cofactors including thiamine (B1), riboflavin (B2), nicotinamide (B3), pantothenate (B5), pyridoxine (B6), biotin (B7), folic acid (B9), and cobalamin (B12). To identify the component(s) in YE that contribute(s) to the repression of cell filamentation upon CapVQ329R overexpression, we supplemented TB medium with these individual B vitamins. While supplementation with thiamine, riboflavin, nicotinamide, pantothenate, biotin, folic acid, and cobalamin showed no effect, supplementation with the vitamin B6 precursor pyridoxine at 5 mg/ml decreased the length of filaments by 50% (Fig. 7a, b and Supplementary Fig. 8). It will be relevant to investigate the molecular mechanism of pyridoxine to inhibit CapVQ329R-induced cell filamentation. Further, the functionality of interconvertible pyridoxal and pyridoxamine vitamin B6 complex compounds and their 5′ phosphate biologically active counterparts can be investigated individually and in combination. In summary, pyridoxine is one component in LB medium, which is effectively inhibiting cell filamentation upon overexpression of CapVQ329R.
The effect of CapVQ329R extends to commensal and UPEC E. coli strains and S. typhimurium
To determine if the effect of CapVQ329R is restricted to E. coli K-12 MG1655, we overexpressed CapVQ329R in commensal and UPEC E. coli strains64,65 and the gastrointestinal pathogen S. typhimurium UMR1. The NCBI database (accessed latest December 15, 2021) indicates CapV homologs to be encoded predominantly by human fecal E. coli strains and strains derived from animals and animal and plant products. UPEC E. coli can develop extended filamentation upon host cell escape20. In all cases, CapVQ329R overexpression inhibited apparent swimming motility (Supplementary Fig. 9a, b).
Furthermore, CapVQ329R induced different degrees of cell filamentation in the E. coli strains and S. typhimurium UMR1 (Supplementary Fig. 9c). CapVQ329R production induced mild cell filamentation in commensal E. coli Fec32 and Fec89, moderate cell filamentation was observed in commensal Fec67 and extensive filamentation occurred in ECOR31, UPEC CFT073, and S. typhimurium UMR1. A heterogeneous population of moderately filamented cells as well as individual rod-shaped cells was observed in the UPEC strain E. coli No. 12 and the commensal E. coli strain Tob1 upon CapVQ329R overexpression (Supplementary Fig. 9c). In summary, our results showed that the phenotypes observed upon CapVQ329R expression were not restricted to E. coli MG1655, but were common to genetically unrelated commensal and UPEC E. coli strains and S. typhimurium, indicating a general effect of CapVQ329R on bacterial cell morphology, regulation of flagella-mediated swimming motility and biofilm formation.
Production of CapVQ329R affects rdar biofilm formation on agar plates
We were subsequently wondering, whether CapVQ329R affects phenotypes other than cell filamentation and flagella expression. To this end, we investigated the colony morphotype on agar plates. A significant number of E. coli isolates display a rdar biofilm morphotype on Congo red agar plates characterized by the expression of the extracellular matrix components amyloid curli and the exopolysaccharide cellulose64.
Congruent with the pronounced temperature-dependent effect of CapVQ329R, we investigated the effect of CapVQ329R in strains that displayed the rdar morphotype at 37 °C. We choose the UPEC strain No. 12, which expresses a semi-constitutive csgD-dependent rdar morphotype65. While overexpression of wild-type CapV had no major effect compared to the No. 12 control, overexpression of CapVQ329R dramatically disrupted the rdar morphotype (Fig. 8a). Observations of colonies by scanning electron microscopy and TEM demonstrated that CapVQ329R induced even more extensive cell filamentation on agar plates compared to liquid culture, without affecting cell arrangement (Fig. 8b). While nonfilamented cells produced a pronounced extracellular matrix, the filaments produced only little or no matrix (Fig. 8b).
a Rdar morphotype in wild-type E. coli No. 12 vector control (VC) and upon overexpression of CapV and its mutant CapVQ329R. Cells were grown on a salt-free LB agar plate for 24 h at 37 °C. b Scanning electron microscopy of plate-grown colonies. Colony morphotypes grown on a salt-free LB agar plate were fixed after 24 h of growth at 37 °C. c CsgD production upon overexpression of CapV and CapVQ329R in E. coli strain No. 12 compared to VC. Only colony morphotypes from the same plate and signals from the same western blot are compared. d LC-MS/MS quantification of in vivo amounts of c-di-GMP, cAMP, and cAMP-GMP upon overexpression of CapV and CapVQ329R in E. coli strain No. 12 compared to VC. Data are displayed as absolute amounts referred to the original cell suspension. VC = pBAD28. pCapV = CapV cloned in pBAD28; Q329R = CapVQ329R cloned in pBAD28.
In conjunction with rdar morphotype downregulation, its major transcriptional activator CsgD was downregulated (Fig. 8c), demonstrating that CapVQ329R expression affects the central regulatory hub for rdar biofilm formation in strain No. 12. Furthermore, CapVQ329R overexpression equally downregulated the rdar morphotype in the commensal E. coli strains ECOR31, Fec67, Fec89, and Tob1 (Supplementary Fig. 9d). Downregulation of CsgD expression upon expression of CapVQ329R was exemplarily observed for strain ECOR31 (Supplementary Fig. 9e).
We then investigated the effect of CapVQ329R overexpression in commensal E. coli MG1655 and UPEC CFT073 which display a smooth and white morphotype when grown at 37 °C, indicative for the lack of rdar biofilm expression under these experimental conditions. After 48 h of growth, though, a distinctly structured brown colony morphology with dye uptake was displayed upon CapVQ329R, but not upon CapV overexpression (Supplementary Fig. 9d). In the same line, S. typhimurium strain UMR1 displayed a smooth and white morphotype at 37 °C66, with its colony to develop a similar brown morphotype after 48 h of CapVQ329R overexpression (Supplementary Fig. 9d). Again, this colony morphotype seem to be distinct from the rdar biofilm in coloration with no or below detection limit production of the biofilm regulator CsgD in E. coli MG1655 (Supplementary Fig. 9). Semi-constitutive rdar morphotype expressing ECOR31 and No. 12 displaying reduced morphology and coloration retained low CsgD levels upon CapVQ329R production (Fig. 8c and Supplementary Fig. 9c). Thus, CapVQ329R production affects biofilm-associated colony morphotype formation in a complex way in different E. coli strains.
CapV Q329R expression alters cyclic (di) nucleotide concentrations
In Enterobacteriaceae, cyclic di-GMP is a ubiquitous bacterial second messenger, which stimulates the development of the rdar biofilm morphotype via csgD expression40,67,68. We investigated the cyclic (di) nucleotide levels exemplarily in the UPEC strain E. coli No. 12. Along with inhibition of csgD expression, the in vivo cyclic di-GMP level was concomitantly decreased upon CapVQ329R overexpression (Fig. 8d). Besides cyclic di-GMP, both cAMP and cAMP-GMP regulate E. coli biofilm formation43,69. Consistent with a downregulated rdar biofilm phenotype, a reduction in cAMP and an increase in cAMP-GMP levels were observed upon CapVQ329R overexpression (Fig. 8d). Thereby, CapVQ329R might, for example, inhibit a diguanylate cyclase or promote phosphodiesterase activity to downregulate the cyclic di-GMP level. Taken together, these results indicate that CapVQ329R inhibits rdar biofilm formation potentially through regulation of the intracellular level of various cyclic (di)-nucleotide signals.
CapVQ329R enhances the susceptibility of E. coli MG1655 to bacteriophage P1 infection and the antibiotic cephalexin
Recently, the patatin-like phospholipase CapV and the dinucleotide cyclase DncV have been shown to take part in bacterial antiphage defense70. Since CapVQ329R showed a physiological function independent of DncV, we wondered whether CapVQ329R still contributes to antiphage defense. While overexpression of wild-type CapV had no effect, interestingly, an approximately tenfold higher plaque formation was observed upon CapVQ329R overexpression compared to the control when MG1655 cells were infected with P1 phage (Supplementary Fig. 10a), indicating that CapVQ329R enhances susceptibility to bacteriophage P1 infection.
We also observed that overexpression of CapVQ329R, but not CapV renders the laboratory strain E. coli MG1655, the commensal strain ECOR31, and UPEC No. 12 more susceptible or sensitive against the beta-lactam antibiotic cephalexin (Supplementary Fig. 10b). A systematic investigation of altered susceptibility to various antibiotic classes awaits to be performed.
We were also wondering whether CapVQ329R-induced cell elongation and concomitant physiological changes impair the interaction of E. coli with host cells. To this end, we exposed the bladder epithelial cell line T24 to UPEC E. coli strain No. 12 overexpressing CapV and CapVQ329R. CapVQ329R expressing E. coli strain No. 12 associated significantly less with the T24 bladder epithelial cell line than the control and CapV expressing cells. Although a trend to lower induction of mRNA steady-state levels of genes coding for the pro-inflammatory markers IL-1β and CXCL-8 was observed for T24 cells exposed to CapVQ329R expressing bacteria, the obtained results were not statistically significant (Supplementary Fig. 10c). Thus, in summary, CapVQ329R production causes substantial physiological alterations in E. coli MG1655.
Discussion
Rod-shaped bacteria can undergo filamentation upon dysregulation of cytokinesis components. In this study, we describe a variant of the patatin-like phospholipase CapV containing a single amino acid substitution, CapVQ329R, to repress the apparent swimming motility and to induce extensive sulA-independent cell filamentation in E. coli MG1655, while only slightly decreasing FtsZ levels. Besides these substantial morphological changes, which occur in both commensal and pathogenic E. coli strains, modulation of rdar biofilm formation occurs concomitantly in the stationary phase of growth. Alteration in local or overall concentrations of cyclic (di)-nucleotides upon CapVQ329R overexpression might be involved in motility and rdar biofilm regulation. Collectively, these findings demonstrate a single amino acid change to create a protein, which dramatically modulates various aspects of bacterial physiology.
CapV of E. coli ECOR31 is a patatin-like PLA2 that contains a N-terminal PNPLA domain and a C-terminal domain of unknown function (Fig. 3b, c). Patatin-like phospholipases proteins are predominantly phospholipase A2 enzymes, acyl hydrolases cleaving the sn-2 position of neutral lipids and phospholipids. We did not observe fundamental alterations in the lipid profile after 4 h of CapVQ329R induction where extensive filamentation had already occurred indicating that CapV and CapVQ329R display only a minor, if any, catalytic activity in the genetic background of E. coli MG1655 (Fig. 3d, e, f and Supplementary Fig. 4). In plants, patatin-like phospholipases do not only act as enzymes to cleave fatty acids from membrane lipids but also aid in the control of the spread of infection. Such a protein is, for example, highly abundant as a storage protein in potato tubers71,72. In mammals, PLPs have been recognized to be involved in lipid metabolism and turnover73, with a polymorphism in a patatin-like phospholipase to provide the genetic predisposition for nonalcoholic fatty liver disease and metabolic syndrome74. Furthermore, a patatin-like phospholipase is involved in axon functionality of neurons75. Patatin-like phospholipases are also found in pathogenic bacterial species and act as toxins in host–pathogen interactions53,76. One of the best characterized patatin-like phosphlipases is ExoU, a cytotoxic effector protein of P. aeruginosa secreted through the type III secretion system upon host cell contact77. Interaction with host ubiquitin and ubiquitinated proteins triggers the PLA2 activity of ExoU leading to degradation of host cell membranes78. Our finding that the G–G-G-x-[K/R]-G and D–G-[A/G] motifs in the PNPLA domain of CapVQ329R, but not the catalytic G-x-S-x-G motif, are required for the cell filamentation phenotype suggests a functional role for the protein scaffold (Fig. 2d). Since bacterial filamentation also contributes to pathogenesis by escaping from phagocytosis, we hypothesize that CapVQ329R has a role in bacterial survival upon induction of cell filamentation during interaction with host immune cells.
The amino acid Q329 is located C-terminal of the putative PNPLA domain of CapV of E. coli ECOR31 and consequently not part of the characteristic motifs required for the catalytic activity of this PLA2 superfamily member (Fig. 3b). 3D model construction using the closest homolog of CapV from E. coli ECOR31 in the PDB database (FabD from Solanum cardiophyllum, PDB: 1oxwC) revealed that Q329 is located in the context of the RARGRR329 sequence motif within the second last α helix of the CapV model structure with the arginine side chain pointing outwards, causing no change in the overall structure (Fig. 3c). However, the switch from a neutral hydrogen-bond acceptor (Q) to a hydrogen-bond donor (R) with a longer side chain indicates that CapVQ329R might show altered enzymatic activity, ligand binding properties or protein-protein interactions. Arginine with its positively charged side chain can be involved in a variety of different functionalities such as in binding of negatively charged phosphates of DNA molecules or nucleotides. The RR twin-arginine motif is part of the N-terminal signal sequence for the Twin-Arginine Translocation (TAT) pathway79,80. Furthermore, a RxxR motif constitutes a conserved peptidase cleavage site, while a RxxxR motif is part of the binding motif for c-di-GMP in PilZ domain proteins, whereby the arginine residues bind O-6 and N-7 at the Hoogsteen edge of the guanine base81.
The catalytic activity of bacterial and eukaryotic patatin-like phospholipases is highly regulated to require a cofactor for activation48. CapV from V. cholerae El Tor is activated by binding to 3′3′-cAMP-GMP synthesized by the dinucleotide cyclase DncV to cause growth retardation46. Up to now, the binding site of cAMP-GMP in CapV awaits identification, while our preliminary experimentation suggests that CapVQ329R and CapV bind cAMP-GMP with equal affinity. Our result showed that the CapV variant CapVQ329R did hardly affect E. coli MG1655 cell viability in the logarithmic growth phase, but induced a 50% growth retardation and loss of cell viability in the stationary phase (Fig. 5 and Supplementary Fig. 7). We cannot exclude cAMP-GMP binding to be required for expression of motility and filamentation phenotypes, but consider this as unlikely. The in vivo cAMP-GMP concentration was below the detection limit for the E. coli K-12 MG1655 strain. Cyclic AMP-GMP could be produced by a GGDEF diguanylate cyclase with altered substrate specificity in the uropathogenic strain No. 12 upregulated upon CapVQ329R expression. Alternatively, c-di-GMP binding could be required for residual stimulation of catalytic activity. As the catalytic serine residue in the G-x-S-x-G motif of the PNPLA domain is not involved in triggering motility inhibition and cell filamentation, we cannot exclude that CapVQ329R might induce the respective phenotypes by residual enzymatic activity, solely through its altered protein scaffold (independent of cAMP-GMP binding) or modification of an enzymatic activity other than the phospholipase activity.
Bacterial cell filamentation is often associated with activation of the SOS response and the cell division inhibitor SulA16,32. However, also mutations in metabolic genes cause filamentation in E. coli19. According to our results, CapVQ329R induced a sulA-independent filamentation in MG1655 (Supplementary Fig. 6d), indicating CapVQ329R affects cell morphology possibly via an alternative signaling pathway.
Besides filamentation, CapVQ329R overexpression inhibited apparent swimming motility of E. coli MG1655 in the soft agar plate (Fig. 1a), with CapVQ329R overproducing cells concomitantly to gradually loose flagella to become non-motile (Supplementary Movie S1). Long filamentous cells, even if observed being motile when up to 20 times the length of a wild-type cell in liquid culture, might become physically trapped in the pores of the agar.
Interestingly, CapVQ329R also downregulated rdar biofilm formation in several E. coli strains that express the morphotype at 37 °C (Supplementary Fig. 9d and Fig. 7a). Examination of cells showed CapVQ329R to remodel colony morphology on the agar plate with the appearance of highly filamented cells along with a light-brown colony morphotype (Fig. 7b). Besides the reduction of in vivo c-di-GMP level, a reduction in cAMP level and a weak signal for cAMP-GMP were detected upon CapVQ329R overexpression in the UPEC E. coli strain No. 12 (Fig. 7d). Cyclic AMP has been shown to promote extracellular matrix production and biofilm formation in UPEC69. Our previous investigation showed that DncV-synthesized cAMP-GMP participates in the downregulation of rdar biofilm formation in E. coli ECOR3143. Besides DncV, a certain class of GGDEF domain proteins has been identified to catalyze the synthesis of cAMP-GMP82. Inspection of the genome sequence did not indicate the presence of a dncV homolog in the E. coli No. 12 genome (unpublished observation), suggesting that cAMP-GMP production by a GGDEF domain protein(s) might be directly or indirectly activated by CapVQ329R, to inhibit rdar biofilm formation of E. coli No. 12. Furthermore, the induction of a csgD independent alternative colony morphotype with distinct dye-binding capability in isolates with a smooth and white morphotype points to a complex regulation of bacterial colony morphology by CapVQ329R.
We show in this work that CapVQ329R expression increased susceptibility to infection by the myophage P1 (Supplementary Fig. 10a). Of note, gene products of the capV-dncV-vc0180-vc0181 operon (Supplementary Fig. 1) of E. coli and V. cholerae in distinct combinations confer immunity against various bacteriophages70. Upon phage infection, the dinucleotide cyclase DncV catalyzes the synthesis of cAMP-GMP which in turn activates the patatin-like phospholipase CapV to eventually abort infection by leading to cell death70. As a mechanism of increased susceptibility, CapVQ329R might alter the abundance of the terminal glucose on core lipopolysaccharide, which serves as the P1 receptor thereby increasing phage-bacterial interaction, enhance the efficiency of DNA injection or manipulate other mechanisms to lead to higher effectivity of P1 infection. Whether the effect of CapVQ329R on phages infection extends to alternative phages needs to be investigated.
In conclusion, our study reported CapVQ329R, a putative patatin-like phospholipase, to inhibit apparent swimming motility and rdar biofilm formation, while to promote extensive cell filamentation, antimicrobial susceptibility and susceptibility to phage P1 (Fig. 9). The major physiological effects extend beyond the laboratory model E. coli K-12 MG1655 to various commensal and clinical E. coli strains and S. typhimurium, demonstrating a single amino acid substitution to evolve protein functionality towards a dramatic modulation of various aspects of bacterial physiology. It remains to be shown whether CapVQ329R can demonstrate physiological effects even beyond these two closely related enterobacteria and whether other phospholipid bilayer-modifying enzymes show a similar effect.
Overexpression of CapVQ329R in E. coli MG1655 causes substantial morphological and physiological changes which lead to cell elongation with extensive filamentation as a consequence, disparate FtsZ-ring formation, impaired nucleoid segregation, and condensation. a change in the overall lipid profile of the cells, downregulation of flagella expression and motility, downregulation of rdar biofilm morphology and production of its activator CsgD and enhanced susceptibility to infection by the myophage P1 and the cell wall inhibiting antibiotic cephalexine. In the clinical isolate E. coli No. 12, reduced adhesion to the bladder epithelial cell line T24 has been observed. In strains not producing the rdar morphotype at 37 °C, a potentially novel biofilm morphotype has been observed. Cephalexin structure from PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/Cephalexin); structure of oleic acid from Wikipedia (https://en.wikipedia.org/wiki/Oleic_acid).
Methods
Bacterial strains and growth conditions
All strains used in this study are listed in Supplementary Table S1. Strains were cultured in tryptone broth (TB, 1% tryptone, 0.5% NaCl), Luria-Bertani (LB, 1% tryptone, 0.5% yeast extract, 1% NaCl) liquid medium or on LB agar plates supplemented with 25 µg/ml chloramphenicol (Cm) at indicated temperatures. l-arabinose (Sigma) was used for induction of gene expression at indicated concentrations.
Plasmid construction
All plasmids and primers used in this study are listed in Supplementary Tables S1–S3, respectively. Genes of interest were amplified by PCR, the PCR products digested with indicated restriction enzymes and ligated into the pBAD28 vector using the Rapid DNA Ligation Kit (Roche Diagnostics). Inserted DNA sequences were confirmed by DNA sequencing (StarSeq).
Site-directed mutagenesis
Site-directed mutagenesis was performed using the Q5 site-directed mutagenesis kit according to the manufacturer’s instructions (NEB). All mutations were confirmed by DNA sequencing.
Swimming assay
To assess apparent swimming motility, 3 µl of an overnight culture suspended in water (OD600 = 5) was inoculated into soft agar medium containing 1% tryptone, 0.5% NaCl, and 0.25% agar83. The swimming halo was documented with a Gel Doc imaging system (Bio-rad) after 6 h at 37 °C and the swimming diameter was measured by ImageJ 1.8.084.
Rdar colony morphotype assessment
To visualize expression of cellulose and curli fimbriae, 5 µl of an overnight culture of OD600 = 5 suspended in water was spotted onto LB without NaCl agar plates containing the dye Congo red (40 µg/ml, Sigma) and Coomassie Brilliant Blue G-250 (20 µg/ml, Sigma). Plates were incubated at 37 °C. Pictures were taken at different time points to analyze the development of the colony morphology structure and dye binding.
Antimicrobial susceptibility assay
Antimicrobial susceptibility was assessed by the disc diffusion assay. Cells were spread on a Mueller Hinton agar plate containing 0.1% arabinose and 25 µg/ml Cm. After 3 h of incubation at 37 °C, discs with 30 µg cephalexin were placed and incubation was continued for another 16 h. Zones of inhibition and sensitivity were assessed visually and by measuring the radius of the zone of inhibition and sensitivity as indicated by no and reduced growth, respectively.
Isolation of cell-associated flagellin
Bacterial cell-associated flagellin was isolated as described previously.85 Briefly, a single bacterial colony of each group was inoculated in TB medium and cultured overnight at 37 °C at 150 rpm. After dilution to OD600 = 0.01, 0.1% l-arabinose was added for induction of the protein, and culturing was continued for indicated time points. Flagella were sheared off by forcing the sample 15 times through a syringe with a needle of 0.51 mm diameter (BD Microlance). One ml of samples was collected, centrifuged at 17,000 rpm and the supernatant was mixed with cold trichloroacetic acid (v:v = 3:1, Sigma). Samples were incubated at −20 °C for 2 h, followed by centrifugation at 17,000 rpm for 40 min at 4 °C. The cell pellet was collected for SDS-PAGE analysis (4% stacking gel, 12% running gel) and the gel was stained by colloidal Coomassie brilliant blue (Sigma). Uncropped gels are exemplarily shown in the Supplementary Information file.
Flagella staining
Staining of bacterial flagella according to Leifson was performed using a flagella stain kit according to the instructions of the manufacturer (BASO Diagnostics). Flagella were observed under a light microscope (Leica MC170 HD).
Transmission electron microscopy
Bacterial flagella were visualized by transmission electron microscopy (TEM)43. Briefly, a suspension of bacterial cells grown overnight was diluted in TB medium with protein expression induced by 0.1% l-arabinose for indicated time points. An aliquot of 3 µl from each sample was added to a grid with a glow discharged carbon-coated supporting film for 3 min. The excess solution was soaked off by a filter paper and the grid was rinsed by adding 5 µl distilled water for 10 s. Distilled water was soaked off again and the grid was stained with 5 µl 1% uranyl acetate (Sigma) in water for 7 sec. Excess stain was soaked off and the grid was air-dried. The samples were examined in a Hitachi HT 7700 (Hitachi, Tokyo, Japan) electron microscope at 80 kV and digital images were taken with a Veleta camera (Olympus, Münster, Germany).
Scanning electron microscopy
The bacterial colonies were exposed to vapor from a filter paper soaked with 4% glutaraldehyde fixed onto the lid of the petri dish immediately after growth. Subsequently, colonies were fixed in 1.5 ml fixative solution (0.5% glutaraldehyde, 2.5% paraformaldehyde in 10 mM HEPES, pH 7.0) for 2 h at 4 °C. The fixed biofilm sample was processed for observation by scanning electron microscopy (SEM) applying dehydration with acetone, critical-point drying, and sputters coating with gold/palladium. Samples were examined in a Zeiss Merlin field emission scanning electron microscope at an acceleration voltage of 5 kV with the Everhart-Thornley SE-detector and the inlens SE-detector in a 30:70, 70:30, or 77:23 ratio.
Light and fluorescence microscopy
Cell morphology was investigated either under the light microscope (Leica MC170 HD) or a fluorescence microscope (Nikon). The viability of cells was analyzed with the LIVE/DEAD BacLight fluorescence stain from Life Technologies (Thermo Fisher Scientific). For all measurements, live cells were mounted onto 1% agarose pads supplemented with TB medium prior to microscopy analysis. Phase-contrast and fluorescence images were captured using a Ti eclipse inverted research microscope (Nikon) with a ×100/1.45 numerical aperture (NA) objective (Nikon). Image processing and cell length measurement was conducted by Fiji ImageJ 1.8.084.
Lipid extraction
Lipid extraction was performed as described previously with slight modifications46,86. Briefly, overnight cultures were inoculated into 50 ml of TB to OD600 = 0.01, expression of gene products induced by 0.1% arabinose, and grown for 4 h at 37 °C. Cells were collected by centrifugation at 4 °C for 20 min, and 5 ml organic extraction buffer (methanol:chloroform:0.1 M formic acid = 20:10:1, v/v/v) was added to the cell pellets, followed by 1 h of heavy shaking at room temperature. 2.5 ml inorganic aqueous buffer (0.2 M H3PO4, 1 M KCl) was added, followed by another 1 h of heavy shaking. The mixture was centrifuged at 13,000×g for 10 min and lipids dissolved in the lower chloroform phase were collected for analysis by mass spectrometry.
Lipidomics analysis
Extracted lipids were analyzed using ultra-high-pressure liquid chromatography (UHPLC) on a Waters CSH column, interfaced to a quadrupole/time-of-flight (QTOF) mass spectrometer (high resolution, accurate mass), with a 15 min total run time. The LC/QTOF MS analyses were performed using an Agilent 1290 Infinity LC system (G4220A binary pump, G4226A autosampler, and G1316C Column Thermostat) coupled to either an Agilent 6530 (positive ion mode) or an Agilent 6550 mass spectrometer equipped with an ion funnel (iFunnel; (negative ion mode). Lipids were separated on an Acquity UPLC CSH C18 column (100 × 2.1 mm; 1.7 µm) maintained at 65 °C at a flow rate of 0.6 ml/min. Solvent pre-heating (Agilent G1316) was used. The mobile phases consisted of 60:40 acetonitrile:water with 10 mM ammonium formate and 0.1% formic acid (A) and 90:10 propan-2-ol:acetonitrile with 10 mM ammonium formate and 0.1% formic acid. The gradient was as follows: 0 min 85% (A); 0–2 min 70% (A); 2–2.5 min 52% (A); 2.5–11 min 18% (A); 11–11.5 min 1% (A); 11.5–12 min 1% (A); 12–12.1 min 85% (A); 12.1–15 min 85% (A).
Samples were injected (1.7 µl in positive mode and 5 µl in negative ion mode) with a needle wash for 20 s (wash solvent is isopropanol). The valve is switched back and forth during the run for washing to reduce carryover of less polar lipids. The sample temperature is maintained at 4 °C in the autosampler.
Data from six samples for each experiment were collected in both positive and negative ion mode, and analyzed using MassHunter (Agilent). Lipids were identified based on their unique MS/MS fragmentation patterns using the software Lipidblast (https://fiehnlab.ucdavis.edu/projects/LipidBlast).87
Principle component analysis was performed on the datasets with GraphPad 9.0 software. The heatmap was constructed on the website https://www.cloudtutu.com/#/login using default parameters with average as the clustering method. In short, values for peak height were log2 transformed, the mean for each lipid species calculated and subsequently each value was mean centered and displayed as a color code.
Abbreviation of lipid compounds as follows: PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PE P, plasmenyl-phosphatidylethanolamine; PI, phosphatidylinositol; PMeOH, phosphatidylmethanol; PS, phosphatidylserine; SM, sphingomyelin; TG, triglyceride; PC P, plasmenyl-phosphatidylcholine; PC O, plasmanyl-phosphatidylcholine; PC, phosphatidylcholine; PA, phosphatidic acid; MGDG, monogalactosyldiacylglycerol; LPG, lysophosphatidylglycerol; LPE, lysophosphatidylethanolamine; LPC O, plasmanyl-lysophosphatidylcholine; LPC, lysophosphatidylcholine; LPA, lysophosphatidic acid; HBMP, 1-Monoacylglycerol-phospho-2,3-diacylglycerol; GlcCer, glycosylceramide; FAHFA, fatty acid ester of hydroxyl fatty acid; FA, fatty acid; DG, diacylglycerol; CL, cardiolipin; Cer, ceramide; CE, cholesteryl ester; SM, sphingomyelin; CAR, acylcarnitine.
Extraction of in vivo-produced nucleotides
Extraction of cyclic dinucleotides from bacterial cells was performed as reported88. Briefly, individual colonies were inoculated in TB medium, overnight cultures diluted to OD600 = 0.01, and grown in TB medium at 37 °C for 4 h containing 0.1% L-arabinose to induce expression of CapV and CapVQ329R. In all, the 5-ml cell suspension was pelleted and resuspended in 500 µl ice-cold extraction solvent (acetonitrile/methanol/water/formic acid = 2/2/1/0.02, v/v/v/v), followed by boiling for 10 min at 95 °C. Three subsequent extracts were combined and frozen at −20 °C overnight. The extracts were centrifuged for 10 min at 20,800×g, evaporated to dryness in a Speed-Vac (Savant), and analyzed by LC-MS/MS89,90. The detection limits for cyclic di-GMP and cAMP were 0.065 and 0.031 pmol/mg wet cell weight, respectively. The detection limits for cAMP-GMP and cyclic di-AMP were 0.235/0.027 and 0.054/0.012 pmol/mg for E. coli strains No. 12/MG1655, respectively.
Western blot analysis
To detect protein expression, 5 mg (wet weight) of bacterial cells collected from agar plates or liquid cultures were resuspended in 200 µl SDS sample buffer and heated at 95 °C for 10 min. The total protein content was assessed by Coomassie Brilliant blue staining after sample separation. Samples containing equal amounts of protein were separated by SDS-PAGE (4% stacking and 12% resolving gel) and transferred onto a PVDF membrane (Millipore). The membrane was blocked with 5% skim milk (for detection of E. coli CsgD and flagellin FliC) or 5% BSA (for detection of His-tagged protein) in blocking buffer overnight. CsgD and FliC were detected with a polyclonal E. coli anti-CsgD peptide primary antibody (dilution 1:5000)91 and anti-Flagellin (FliC) antibody (ab93713, Abcam, dilution 1:5000), respectively. The horseradish peroxidase-conjugated goat anti-rabbit IgG was the secondary antibody (154-10004301, Jackson ImmunoResearch Laboratories Inc., dilution 1:2000). An anti·His antibody conjugated to horseradish peroxidase (Penta·His HRP Conjugate, ID: 34460, Qiagen, dilution 1:2000) was used to detect 6xHis-tagged proteins. Antibody binding was visualized with ECL light detection reagent (Roche) using Luminescent Image Analyzer (LAS-1000plus, Fujifilm). Uncropped blots are exemplarily shown in the Supplementary Information file.
Total RNA isolation and quantitative real-time PCR (qPCR) of bacterial cells
Bacterial cells were grown in TB medium for 6 h with CapV/CapVQ329R expression induced from the pBAD28 vector. Total RNA was isolated by hot acidic phenol extraction as previously described92 with subsequent assessment of RNA concentration by recording an absorbance spectrum from 220 to 350 nm by the NanoDrop 2000 (Thermo Scientific) and assessment of RNA integrity and DNA contamination upon separation in a 1% agarose gel run with 0.5 × TBE buffer. In total, 1 µg of RNA was used for cDNA synthesis by the High-Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was run in the ABI 7500 real-time PCR system (Applied Biosystems) using the RT-qPCR ReadyMixTM (Sigma) with primers indicated in Supplementary Table S4. Reference genes were recA and rspV. The reference for relative target gene expression was the expression in the vector control strain calculated as 2−∆∆CT.
Phage P1 plaque-formation assays
The phage P1 plaque-formation assay was performed according to previously described methods with slight modifications70,93. Individual bacterial colonies from overnight cultures were inoculated into TB medium and grown to OD600 = 0.1 at 37 °C, followed by further growth with 0.1% l-arabinose for another 3 h to induce the expression of CapV and CapVQ329R. For the determination of phage infectivity by plaque formation, 500 µl of cells were thoroughly mixed with 4.5 mL modified MMB agar (TB with 0.1 mM MnCl2, 5 mM MgCl2, 0.35% agar), immediately poured onto Petri dishes containing 20 ml MMB Agar (1.6%) and allowed to cool for 10 min at room temperature. Tenfold serial dilutions of P1 phage lysate in SM Buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl pH 7.5) were dropped on top of the double-layer agar plate and allowed to dry for 20 min at room temperature. Plates were incubated at 37 °C for 18 h and plaques were counted to compare the efficiency of plating.
Bioinformatic analyses
A BLAST search against the NCBI protein database was performed with standard parameters using the E. coli ECOR31 CapV sequence as a query. All distinct protein sequences with >40% identity from different species were selected. Sequences were aligned using CLUSTALW94 and processed with ESPript 3.095 using standard parameters. The phylogenetic tree was reconstructed by calculating sequence similarity by Maximum Likelihood (ML) in MEGA 7.096. A CapV and CapVQ329R structural model was built with the I-TASSER server97 and processed with SWISS-MODEL98. Sequences used for alignment and in the phylogenetic tree are: STM4563_SALTY: S. typhimurium NP_463419.1, YjjU_ECOLI: E. coli BAE78366.1, RssA_ECOLI: E. coli NP_415750.2, STM1754_SALTY: S. typhimurium YchK NC_003197.2, PA1640_PSEAI: Pseudomonas aeruginosa SG17M Homolog_EWH27047.1, CapV_ECOR31: E. coli ECOR31 OII97420.1, CapV_VIBCH: V. cholerae NC_002505.1, PLP2_SOLTU: Solanum tuberosum, PA3241_PSEAI: P. aeruginosa SG17M Homolog_EWH29020.1, PatA_LEGPH: Legionella lpg2317 NC_002942.5, RssA_HALMT: Haloferax mediterranei WP_004060664.1, Q68WK1_RICTY: Rickettsia typhi RT0522 WP_011190972.1, VpdB_LEGPH: Legionella pneumophila QGK66366.1, ExoU_PSAER: P. aeruginosa WP_003134060.1, VpdC_LEGPH: L. pneumophila QGK66518.1, VpdA_LEGPH: L. pneumophila CCD09784.1, VipD_LEGPH: L. pneumophila WP_010948518.1, CapE_ECOR31: E. coli ECOR31 OII97423.1, COTR_BACSU: Bacillus subtilis WP_003243674.1.
Host cell adhesion assay-normalized values
Bacterial adhesion was assessed with the human uroepithelial cell line T24 cultured in McCoy’s 5a medium supplemented with 10% fetal bovine serum. E. coli No. 12 harboring pCapV, pCapVQ329R, or vector control pBAD28 were plated on Luria-Bertani agar plates and incubated at 37 °C overnight. The following day, one colony was inoculated in LB with 100 µg/ml of ampicillin and incubated overnight at 37 °C with 200 RPM. For induction of filaments, the cell suspension was diluted 10 times and subcultured in TB broth supplemented with 0.1% l-arabinose and 100 µg/ml of ampicillin at 200 RPM for 2 h. Microscopic observation confirmed filament formation. Infection of T24 host cells was carried out in 24-well cell culture plates (Costar) with 2 × 106 CFU/ml (MOI 10) of E. coli No. 12 harboring pCapV, pCapVQ329R, or the vector control pBAD28 to be added to the cell culture in serum- and antibiotic-free medium and incubated at 37 °C. After 30 min, cells were washed three times with 37 °C warm PBS to remove non-adherent bacteria. To collect cell-associated bacteria, ice-cold PBS with 1% Triton X-100 was added. Lysates were plated on blood agar plates after serial dilution in PBS and bacterial numbers were counted after overnight incubation at 37 °C. The relative adhesion of E. coli No. 12 with pCapV or pCapVQ329R was calculated in relation to vector control.
Total RNA isolation and quantitative real-time PCR (qPCR) of host cells
The human uroepithelial cell line T24 was infected by co-incubation with isogenic strains of E. coli for 2 h99. Total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. The concentration and purity of RNA were determined with nanodrop, and 300 ng of RNA was transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR for human IL1B (forward 5′-CAC GAT GCA CCT GTA CGA TCA-3′, reverse 5′-GTT GCT CCA TAT CCT GTC CCT-3′), CXCL8 (forward 5′-AAG AGA GCT CTG TCT GGA CC-3′, reverse 5′-GAT ATT CTC TTG GCC CTT GG-3′) were analyzed in comparison to housekeeping gene ACTB (forward 5′- AAG AGA GGC ATC CTC ACC CT-3′, reverse 5′-TAC ATC GCT GGG GTG TTG-3′) using SYBR green gene expression assay (Applied Biosystems) in a Rotor-Gene PCR cycler (Corbett Life Science). Relative expressions of target genes was presented as 2−∆CT and fold change as 2−∆∆CT compared to vector control infected cells.
DRaCALA assay
Bacterial cells were prepared for the DRaCALA assay basically as described100. Cells were cultured overnight and subcultured to a starting OD600 of 0.02 induced with 0.1% arabinose. At OD600 = 2, cells were collected resuspended in 10 mM Tris, pH 8.0, 100 mM NaCl, and 5 mM MgCl2. Cells were lysed three times by freeze-thaw and the lysate was tested for binding 32P labelled cAMP-GMP starting with a fourfold dilution. 32P-cAMP-GMP had been prepared by mixing 10 µCi of 32P-α-GTP (8 nmol) and 250 µM ATP followed by addition of 10 µM DncV in reaction buffer (50 mM 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (CAPSO) pH 9.4, 175 mM KCl, 25 mM, 175 mM Mg(OAc)2, and 5 mM DTT) and incubated for 2 h at 37 °C.
Data availability
The data underlying this article are available in the article and in its online Supplementary Information file. Material such as bacterial strains and plasmids related to this paper may be requested from the corresponding author (Ute.Romling@ki.se) or from Addgene under the ID:80770. Lipidomics data have been made available at the Center for Open Science webpage under https://osf.io/aj58e/.
References
Cabeen, M. T. & Jacobs-Wagner, C. Bacterial cell shape. Nat. Rev. Microbiol. 3, 601–610 (2005).
Jones, T. H., Vail, K. M. & McMullen, L. M. Filament formation by foodborne bacteria under sublethal stress. Int. J. Food Microbiol. 165, 97–110 (2013).
Utsumi, R., Nakamoto, Y., Kawamukai, M., Himeno, M. & Komano, T. Involvement of cyclic AMP and its receptor protein in filamentation of an Escherichia coli fic mutant. J. Bacteriol. 151, 807–812 (1982).
Justice, S. S. et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl Acad. Sci. USA 101, 1333–1338 (2004).
Heinrich, K., Leslie, D. J., Morlock, M., Bertilsson, S. & Jonas, K. Molecular basis and ecological relevance of Caulobacter cell filamentation in freshwater habitats. mBio 10, e01557–01519 (2019).
Wortinger, M. A., Quardokus, E. M. & Brun, Y. V. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Mol. Microbiol. 29, 963–973 (1998).
Davis, C. P. & Savage, D. C. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 10, 948–956 (1974).
Wagner, M. et al. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60, 792–800 (1994).
Angert, E. R., Clements, K. D. & Pace, N. R. The largest bacterium. Nature 362, 239–241 (1993).
Pine, L. & Boone, C. J. Comparative cell wall analyses of morphological forms within the genus Actinomyces. J. Bacteriol. 94, 875–883 (1967).
Chen, K., Sun, G. W., Chua, K. L. & Gan, Y.-H. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrob. Agents Chemother. 49, 1002–1009 (2005).
Cushnie, T. T., O’Driscoll, N. H. & Lamb, A. J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 73, 4471–4492 (2016).
Spratt, B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl Acad. Sci. USA 72, 2999–3003 (1975).
Ultee, E., Ramijan, K., Dame, R. T., Briegel, A. & Claessen, D. Stress-induced adaptive morphogenesis in bacteria. Adv. Microb. Physiol. 74, 97–141 (2019).
Radman, M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. in Molecular Mechanisms for Repair of DNA (eds Hanawalt, P. C. & Setlow, R. B.) 355–367 (Springer, 1975).
Cordell, S. C., Robinson, E. J. & Löwe, J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl Acad. Sci. USA 100, 7889–7894 (2003).
Kuhn, I., Peng, L., Bedu, S. & Zhang, C.-C. Developmental regulation of the cell division protein FtsZ in Anabaena sp. strain PCC 7120, a cyanobacterium capable of terminal differentiation. J. Bacteriol. 182, 4640–4643 (2000).
Addinall, S. G., Bi, E. & Lutkenhaus, J. FtsZ ring formation in fts mutants. J. Bacteriol. 178, 3877–3884 (1996).
El-Hajj, Z. W. & Newman, E. B. How much territory can a single E. coli cell control? Front. Microbiol. 6, 309 (2015).
Justice, S. S., Hunstad, D. A., Seed, P. C. & Hultgren, S. J. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl Acad. Sci. USA 103, 19884–19889 (2006).
Jones, C. & Holland, I. Inactivation of essential division genes, ftsA, ftsZ, suppresses mutations at sfiB, a locus mediating division inhibition during the SOS response in E. coli. EMBO J. 3, 1181–1186 (1984).
Balasubramanian, A., Markovski, M., Hoskins, J. R., Doyle, S. M. & Wickner, S. Hsp90 of E. coli modulates assembly of FtsZ, the bacterial tubulin homolog. Proc. Natl Acad. Sci. USA 116, 12285–12294 (2019).
Yadavalli, S. S. et al. Antimicrobial peptides trigger a division block in Escherichia coli through stimulation of a signalling system. Nat. Commun. 7, 1–10 (2016).
Möller, J., Emge, P., Vizcarra, I. A., Kollmannsberger, P. & Vogel, V. Bacterial filamentation accelerates colonization of adhesive spots embedded in biopassive surfaces. N. J. Phy. 15, 125016 (2013).
Miller, C. et al. SOS response induction by ß-lactams and bacterial defense against antibiotic lethality. Science 305, 1629–1631 (2004).
Möller, J., Luehmann, T., Hall, H. & Vogel, V. The race to the pole: how high-aspect ratio shape and heterogeneous environments limit phagocytosis of filamentous Escherichia coli bacteria by macrophages. Nano Lett. 12, 2901–2905 (2012).
Bernhardt, T. G. & de Boer, P. A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 (2005).
Hiraga, S. Chromosome partitioning in Escherichia coli. Curr. Opin. Genet. Dev. 5, 789–801 (1993).
Margolin, W. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24, 531–548 (2000).
Lu, C.-H. et al. Fic proteins inhibit the activity of topoisomerase IV by AMPylation in diverse bacteria. Front. Microbiol. 11, 2084 (2020).
Lau, S. Y. & Zgurskaya, H. I. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J. Bacteriol. 187, 7815–7825 (2005).
Bi, E. & Lutkenhaus, J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175, 1118–1125 (1993).
Monteiro, C., Fang, X., Ahmad, I., Gomelsky, M. & Römling, U. Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J. Bacteriol. 193, 6443–6451 (2011).
Raskin, D. M. & de Boer, P. A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181, 6419–6424 (1999).
Khandige, S. et al. DamX controls reversible cell morphology switching in uropathogenic Escherichia coli. mBio 7, e00642–00616 (2016).
Harshey, R. M. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273 (2003).
Park, S.-Y., Pontes, M. H. & Groisman, E. A. Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc. Natl Acad. Sci. USA 112, 1850–1855 (2015).
Wolfe, A. J. & Visick, K. L. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190, 463–475 (2008).
Terashima, H., Kojima, S. & Homma, M. Flagellar motility in bacteria: structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270, 39–85 (2008).
Römling, U., Galperin, M. Y. & Gomelsky, M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013).
Römling, U. Cyclic di‐GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14, 1817–1829 (2012).
Davies, B. W., Bogard, R. W., Young, T. S. & Mekalanos, J. J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012).
Li, F. et al. DncV synthesizes cyclic GMP-AMP and regulates biofilm formation and motility in Escherichia coli ECOR31. mBio 10, e02492–02418 (2019).
Schubert, S., Dufke, S., Sorsa, J. & Heesemann, J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high‐pathogenicity island. Mol. Microbiol. 51, 837–848 (2004).
Whiteley, A. T. et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019).
Severin, G. B. et al. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc. Natl Acad. Sci. USA 115, E6048–E6055 (2018).
Cohen, D. et al. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019).
Scherer, G. F., Ryu, S. B., Wang, X., Matos, A. R. & Heitz, T. Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700 (2010).
Kato, K., Ishii, R., Hirano, S., Ishitani, R. & Nureki, O. Structural basis for the catalytic mechanism of DncV, bacterial homolog of cyclic GMP-AMP synthase. Structure 23, 843–850 (2015).
Ryjenkov, D. A., Simm, R., Römling, U. & Gomelsky, M. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314 (2006).
Sigrist, C. J. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 (2012).
Six, D. A. & Dennis, E. A. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1488, 1–19 (2000).
Banerji, S. & Flieger, A. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150, 522–525 (2004).
Lutkenhaus, J. FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9, 403–409 (1993).
Thanbichler, M. Synchronization of chromosome dynamics and cell division in bacteria. Cold Spring Harb. Perspect. Biol. 2, a000331 (2010).
Sherratt, D. J. Bacterial chromosome dynamics. Science 301, 780–785 (2003).
Söderström, B. et al. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol. Microbiol. 92, 1–9 (2014).
Ma, X., Ehrhardt, D. W. & Margolin, W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA 93, 12998–13003 (1996).
Harms, A., Fino, C., Sørensen, M. A., Semsey, S. & Gerdes, K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8, e01964–01917 (2017).
Goormaghtigh, F. & Van Melderen, L. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci. Adv. 5, eaav9462 (2019).
Bisicchia, P., Arumugam, S., Schwille, P. & Sherratt, D. MinC, MinD, and MinE drive counter-oscillation of early-cell-division proteins prior to Escherichia coli septum formation. mBio 4, e00856-13 (2013).
Zarei, O., Dastmalchi, S. & Hamzeh-Mivehroud, M. A simple and rapid protocol for producing yeast extract from Saccharomyces cerevisiae suitable for preparing bacterial culture media. Iran J. Pharm. Res. 15, 907–913 (2016).
Walker, G. M. Yeast Physiology and Biotechnology (John Wiley & Sons, 1998).
Bokranz, W., Wang, X., Tschäpe, H. & Römling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54, 1171–1182 (2005).
Cimdins, A. et al. Alterations of c‐di‐GMP turnover proteins modulate semi‐constitutive rdar biofilm formation in commensal and uropathogenic Escherichia coli. MicrobiologyOpen 6, e00508 (2017).
Römling, U., Sierralta, W. D., Eriksson, K. & Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28, 249–264 (1998).
Römling, U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62, 1234–1246 (2005).
Hammar, M., Arnqvist, A., Bian, Z., Olsén, A. & Normark, S. Expression of two csg operons is required for production of fibronectin‐and congo red‐binding curli polymers in Escherichia coli K‐12. Mol. Microbiol. 18, 661–670 (1995).
Hufnagel, D. A. et al. The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J. Bacteriol. 198, 3329–3334 (2016).
Cohen, D. et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019).
Rosahl, S., Schmidt, R., Schell, J. & Willmitzer, L. Isolation and characterization of a gene from Solanum tuberosum encoding patatin, the major storage protein of potato tubers. Mol. Gen. Genet. MGG 203, 214–220 (1986).
Andrews, D., Beames, B., Summers, M. & Park, W. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J. 252, 199–206 (1988).
Wilson, S. K. & Knoll, L. J. Patatin‐like phospholipases in microbial infections with emerging roles in fatty acid metabolism and immune regulation by Apicomplexa. Mol. Microbiol. 107, 34–46 (2018).
Bruschi, F. V., Tardelli, M., Claudel, T. & Trauner, M. PNPLA3 expression and its impact on the liver: current perspectives. Hepat. Med. 9, 55–66 (2017).
Dutta, S., Rieche, F., Eckl, N., Duch, C. & Kretzschmar, D. Glial expression of Swiss cheese (SWS), the Drosophila orthologue of neuropathy target esterase (NTE), is required for neuronal ensheathment and function. Dis. Models Mech. 9, 283–294 (2016).
Tessmer, M. H., Anderson, D. M., Pickrum, A. M., Riegert, M. O. & Frank, D. W. Identification and verification of ubiquitin-activated bacterial phospholipases. J. Bacteriol. 201, e00623–00618 (2019).
Sawa, T. et al. Pseudomonas aeruginosa type III secretory toxin ExoU and its predicted homologs. Toxins 8, 307 (2016).
Anderson, D. M. et al. Ubiquitin and ubiquitin‐modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 82, 1454–1467 (2011).
Frain, K. M., Dijl, J. M. V. & Robinson, C. The twin‐arginine pathway for protein secretion. in Protein Secretion in Bacteria (eds Sandkvist, M., Cascales, E. & Christie, P. J.) 53–66 (ASM Press, 2019).
Lin, B.-L. et al. The arginine pairs and C-termini of the Sso7c4 from Sulfolobus solfataricus participate in binding and bending DNA. PLoS ONE 12, e0169627 (2017).
Galperin, M. Y. & Chou, S.-H. Structural conservation and diversity of PilZ-related domains. J. Bacteriol. 202, e00664–19 (2020).
Hallberg, Z. F. et al. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′, 3′-cGAMP). Proc. Natl Acad. Sci. USA 113, 1790–1795 (2016).
Fang, X. & Gomelsky, M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Le Guyon, S., Rhen, M. & Römling, U. Rapid preparation of unsheathed bacterial flagella. BioProtoc. 5, e1425 (2015).
Wang, Z. & Benning, C. Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 18, e2518 (2011).
Kind, T., Liu, K.H., Lee, D.Y., DeFelice, B., Meissen, J.K. & Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 10, 755–758 (2013).
Burhenne, H. & Kaever, V. Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. In Cyclic Nucleotide Signaling in Plants (ed. Gehring, C.) 27–37 (Humana Press, 2013).
Liu, Y. et al. A cyclic di-GMP network is present in Gram-positive Streptococcus and Gram-negative Proteus species. ACS Infect. Dis. 6, 2672–2687 (2020).
Bähre, H. & Kaever, V. Identification and quantification of cyclic di-guanosine monophosphate and its linear metabolites by reversed-phase LC-MS/MS. Methods Mol. Biol. 1657, 45–58 (2017).
Monteiro, C. et al. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ. Microbiol. 11, 1105–1116 (2009).
Römling, U., Bian, Z., Hammar, M., Sierralta, W. D. & Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731 (1998).
Lowey, B. et al. CBASS immunity uses CARF-related effectors to sense 3′-5′- and 2′-5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182, 38–49.e17 (2020).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Yang, J. & Zhang, Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Kai-Larsen, Y. et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 6, e1001010 (2010).
Roelofs, K. G. et al. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog. 11, e1005232 (2015).
Acknowledgements
We thank the following individuals for providing strains: Prof. Ken Gerdes (Newcastle University), Prof. Van Melderen (Université Libre de Bruxelles), Prof. Daniel O. Daley (Stockholm University), Prof. David Sherrat (University of Oxford), and Prof. Sun Nyunt Wai (Umeå University). We thank Prof. Jan-Willem de Gier (Stockholm University) for providing phage P1. We are also grateful to Dr. Sulman Shafeeq for support; Aysha Qureshi and Elsa Malmström for experimental support during their internship, and the EM core facility at Karolinska Institutet for processing our samples. Further, we appreciate the help of Ina Schleicher in assisting in the preparation of the scanning electron microscopic samples. We also appreciate the support of Prof. Oliver Fiehn and his team at the University of California in Davis in expert lipidomic analyses. F.L. has received a scholarship from the China Scholarship Council (CSC No. 201406170052)). This work was supported by a grant from the Swedish Research Council for Natural Sciences and Engineering (621-2013-4809) to U.R. and grants from Stiftelsen Olle Engkvist Byggmästare and Region Stockholm (ALF Project) to A.B.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Contributions
F.L., V.T.L., and U.R. conceived the study and designed the experiments. F.L., L.C., H.B., S.M., S.K., S.M., K.K., and M.R. performed the experiments. F.L., H.B., K.S., K.J., F.B., A.B., V.T.L., M.R., and U.R. analyzed the data. F.L. and U.R. wrote the paper. All authors reviewed, revised, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, F., Cao, L., Bähre, H. et al. Patatin-like phospholipase CapV in Escherichia coli - morphological and physiological effects of one amino acid substitution. npj Biofilms Microbiomes 8, 39 (2022). https://doi.org/10.1038/s41522-022-00294-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-022-00294-z
This article is cited by
-
Identification and expression profiling of c-di-GMP signaling genes in the probiotic strain Escherichia coli Nissle 1917 during adhesion to the intestinal epithelial cells
Systems Microbiology and Biomanufacturing (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.