Abstract
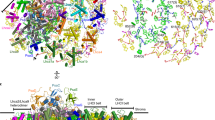
The heart of oxygenic photosynthesis is the water-splitting photosystem II (PSII), which forms supercomplexes with a variable amount of peripheral trimeric light-harvesting complexes (LHCII). Our knowledge of the structure of green plant PSII supercomplex is based on findings obtained from several representatives of green algae and flowering plants; however, data from a non-flowering plant are currently missing. Here we report a cryo-electron microscopy structure of PSII supercomplex from spruce, a representative of non-flowering land plants, at 2.8 Å resolution. Compared with flowering plants, PSII supercomplex in spruce contains an additional Ycf12 subunit, Lhcb4 protein is replaced by Lhcb8, and trimeric LHCII is present as a homotrimer of Lhcb1. Unexpectedly, we have found α-tocopherol (α-Toc)/α-tocopherolquinone (α-TQ) at the boundary between the LHCII trimer and the inner antenna CP43. The molecule of α-Toc/α-TQ is located close to chlorophyll a614 of one of the Lhcb1 proteins and its chromanol/quinone head is exposed to the thylakoid lumen. The position of α-Toc in PSII supercomplex makes it an ideal candidate for the sensor of excessive light, as α-Toc can be oxidized to α-TQ by high-light-induced singlet oxygen at low lumenal pH. The molecule of α-TQ appears to shift slightly into the PSII supercomplex, which could trigger important structure–functional modifications in PSII supercomplex. Inspection of the previously reported cryo-electron microscopy maps of PSII supercomplexes indicates that α-Toc/α-TQ can be present at the same site also in PSII supercomplexes from flowering plants, but its identification in the previous studies has been hindered by insufficient resolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD035272. The cryo-EM map of the spruce PSII supercomplex has been deposited in the Electron Microscopy Data Bank with accession code EMD-16389. The corresponding structure model has been deposited in the PDB under PDB code 8C29.
References
Shen, L. et al. Structure of a C2S2M2N2-type PSII–LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 116, 21246–21255 (2019).
Sheng, X. et al. Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 5, 1320–1330 (2019).
Wei, X. et al. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016).
van Bezouwen, L. S. et al. Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat. Plants 3, 17080 (2017).
Su, X. et al. Structure and assembly mechanism of plant C2S2M2-type PSII–LHCII supercomplex. Science 357, 815–820 (2017).
Kamiya, N. & Shen, J.-R. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl Acad. Sci. USA 100, 98–103 (2003).
Ferreira, K. N., Iverson, T. M., Maghlaoui, K., Barber, J. & Iwata, S. Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 (2004).
Loll, B., Kern, J., Saenger, W., Zouni, A. & Biesiadka, J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 (2005).
Minagawa, J. & Takahashi, Y. Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82, 241–263 (2004).
Jansson, S. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184, 1–19 (1994).
Kouřil, R., Nosek, L., Semchonok, D., Boekema, E. J. & Ilík, P. Organization of plant photosystem II and photosystem I supercomplexes. Subcell. Biochem. 87, 259–286 (2018).
Cao, P., Pan, X., Su, X., Liu, Z. & Li, M. Assembly of eukaryotic photosystem II with diverse light-harvesting antennas. Curr. Opin. Struct. Biol. 63, 49–57 (2020).
Croce, R. & van Amerongen, H. Light harvesting in oxygenic photosynthesis: structural biology meets spectroscopy. Science 369, eaay2058 (2020).
Kouřil, R., Nosek, L., Bartoš, J., Boekema, E. J. & Ilík, P. Evolutionary loss of light-harvesting proteins Lhcb6 and Lhcb3 in major land plant groups—break-up of current dogma. N. Phytol. 210, 808–814 (2016).
Kouřil, R. et al. Unique organization of photosystem II supercomplexes and megacomplexes in Norway spruce. Plant J. 104, 215–225 (2020).
Grebe, S. et al. The unique photosynthetic apparatus of Pinaceae: analysis of photosynthetic complexes in Picea abies. J. Exp. Bot. 70, 3211–3225 (2019).
Klimmek, F., Sjödin, A., Noutsos, C., Leister, D. & Jansson, S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140, 793–804 (2006).
Albanese, P. et al. Dynamic reorganization of photosystem II supercomplexes in response to variations in light intensities. Biochim. Biophys. Acta Bioenerg. 1857, 1651–1660 (2016).
Grinzato, A. et al. High-light versus low-light: effects on paired photosystem II supercomplex structural rearrangement in pea plants. Int. J. Mol. Sci. 21, 8643 (2020).
Kashino, Y. et al. Ycf12 is a core subunit in the photosystem II complex. Biochim. Biophys. Acta 1767, 1269–1275 (2007).
Crepin, A. & Caffarri, S. Functions and evolution of Lhcb isoforms composing LHCII, the major light harvesting complex of photosystem II of green eukaryotic organisms. Curr. Protein Pept. Sci. 19, 699–713 (2018).
Guardini, Z., Gomez, R. L., Caferri, R., Dall’Osto, L. & Bassi, R. Loss of a single chlorophyll in CP29 triggers re-organization of the photosystem II supramolecular assembly. Biochim. Biophys. Acta Bioenerg. 1863, 148555 (2022).
Kruk, J. & Srzałka, K. Occurrence and function of alpha-tocopherol quinone in plants. J. Plant Physiol. 145, 405–409 (1995).
Kumar, A., Prasad, A. & Pospíšil, P. Formation of α-tocopherol hydroperoxide and α-tocopheroxyl radical: relevance for photooxidative stress in Arabidopsis. Sci. Rep. 10, 19646 (2020).
Graça, A. T., Hall, M., Persson, K. & Schröder, W. P. High-resolution model of Arabidopsis photosystem II reveals the structural consequences of digitonin-extraction. Sci. Rep. 11, 15534 (2021).
Triantaphylides, C. & Havaux, M. Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 14, 219–228 (2009).
Krieger-Liszkay, A. & Trebst, A. Tocopherol is the scavenger of singlet oxygen produced by the triplet states of chlorophyll in the PSII reaction centre. J. Exp. Bot. 57, 1677–1684 (2006).
Shi, L. X., Lorkovic, Z. J., Oelmuller, R. & Schroder, W. P. The low molecular mass PsbW protein is involved in the stabilization of the dimeric photosystem II complex in Arabidopsis thaliana. J. Biol. Chem. 275, 37945–37950 (2000).
Granvogl, B., Zoryan, M., Plöscher, M. & Eichacker, L. A. Localization of 13 one-helix integral membrane proteins in photosystem II subcomplexes. Anal. Biochem. 383, 279–288 (2008).
Kruk, J., Schmid, G. H. & Strzałka, K. Interaction of α-tocopherol quinone, α-tocopherol and other prenyllipids with photosystem II. Plant Physiol. Biochem. 38, 271–277 (2000).
Bielczynski, L. W., Xu, P. & Croce, R. PSII supercomplex disassembly is not needed for the induction of energy quenching (qE). Photosynth. Res. 152, 275–281 (2022).
Novoderezhkin, V., Marin, A. & van Grondelle, R. Intra- and inter-monomeric transfers in the light harvesting LHCII complex: the Redfield–Förster picture. Phys. Chem. Chem. Phys. 13, 17093–17103 (2011).
Papadatos, S., Charalambous, A. C. & Daskalakis, V. A pathway for protective quenching in antenna proteins of photosystem II. Sci. Rep. 7, 2523 (2017).
Hakala-Yatkin, M. et al. Magnetic field protects plants against high light by slowing down production of singlet oxygen. Physiol. Plant. 42, 26–34 (2011).
Niewiadomska, E. et al. Lack of tocopherols influences the PSII antenna and the functioning of photosystems under low light. J. Plant Physiol. 223, 57–64 (2018).
Dau, H. et al. Structural consequences of ammonia binding to the manganese center of the photosynthetic oxygen-evolving complex: an X-ray absorption spectroscopy study of isotropic and oriented photosystem II particles. Biochemistry 34, 5274–5287 (1995).
Lichtenthaler, H. K. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382 (1987).
Caffari, S., Kouril, R., Kereiche, S., Boekema, E. J. & Croce, R. Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 (2009).
Sorzano, C. O. S. et al. A new algorithm for high-resolution reconstruction of single particles by electron microscopy. J. Struct. Biol. 204, 329–337 (2018).
Zheng, A. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 4, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
de la Rosa Tevín, J. M. et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 (2013).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Acknowledgements
This work was supported by the Grant Agency of the Czech Republic (project no. 21-05497S to M.O., P.I., P.P., I.I. and R.K.) and the European Regional Development Fund (ERDF) project ‘Plants as a tool for sustainable global development’ (no. CZ.02.1.01/0.0/0.0/16_019/0000827 to M.O., P.I., P.P., R.K., S.Ć.Z. and P.T.). We acknowledge support by the Federal Ministry for Education and Research (BMBF, ZIK programme) (grant nos. 03Z22HN23, 03Z22HI2 and 03COV04 to P.L.K.), Horizon Europe ERA Chair ‘hot4cryo’ project number 101086665 to P.L.K., the European Regional Development Funds for Saxony-Anhalt (grant no. EFRE: ZS/2016/04/78115 to P.L.K.), funding by the Deutsche Forschungsgemeinschaft (DFG) (project number 391498659 and RTG 2467 to P.L.K.), and the Martin-Luther University of Halle-Wittenberg. This work was also funded by project no. RO0423 to S.Ć.Z. and P.T. (Sustainable systems and technologies, improving crop production for higher quality of production of food, feed, and raw materials, under conditions of changing climate) funded by the Ministry of Agriculture, Czechia. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2023042 and European Regional Development Fund-Project ‘UP CIISB’ (no. CZ.02.1.01/0.0/0.0/18_046/0015974), is gratefully acknowledged for the financial support of the measurements at the CEITEC Proteomics Core Facility. We thank L. Hloušková and J. Bartoš for help with FRET rate calculation. This paper is dedicated to Professor Emeritus Jan Nauš for his outstanding contribution to the development of biophysics at Palacký University.
Author information
Authors and Affiliations
Contributions
M.O., D.K., P.I., P.P, P.L.K. and R.K., study design. M.O., D.A.S., F.L.K. and F.H., sample preparation for cryo-EM. D.A.S., image analysis of cryo-EM data. D.K. and R.K., model building. M.O. and I.I., amino acid sequence analysis. P.R., mass spectrometry analysis. P.T. and S.Ć.Z., fatty acid composition. P.T. and S.Ć.Z., α-tocopherol(quinone) analysis. M.O., D.K., P.I., P.P. I.I. and R.K., data interpretation. M.O., D.A.S., P.I., I.I. and R.K. wrote the main body of the paper, and all authors revised and approved it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Jian-Ren Shen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
A description of MS analysis and Supplementary Figs. 1–14, Tables 1–6 and References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Opatíková, M., Semchonok, D.A., Kopečný, D. et al. Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and α-tocopherol. Nat. Plants 9, 1359–1369 (2023). https://doi.org/10.1038/s41477-023-01483-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01483-0