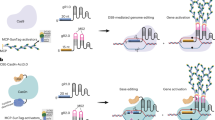
Abstract
CRISPR-Cas9, its derived base editors and CRISPR activation systems have greatly aided genome engineering in plants. However, these systems are mostly used separately, leaving their combinational potential largely untapped. Here we develop a versatile CRISPR-Combo platform, based on a single Cas9 protein, for simultaneous genome editing (targeted mutagenesis or base editing) and gene activation in plants. We showcase the powerful applications of CRISPR-Combo for boosting plant genome editing. First, CRISPR-Combo is used to shorten the plant life cycle and reduce the efforts in screening transgene-free genome-edited plants by activation of a florigen gene in Arabidopsis. Next, we demonstrate accelerated regeneration and propagation of genome-edited plants by activation of morphogenic genes in poplar. Furthermore, we apply CRISPR-Combo to achieve rice regeneration without exogenous plant hormones, which is established as a new method to predominately enrich heritable targeted mutations. In conclusion, CRISPR-Combo is a versatile genome engineering tool with promising applications in crop breeding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All generated and processed data from this study are included in the published article and its Supplementary Information. The Golden Gate and Gateway compatible vectors for the CRISPR-Combo systems were deposited to Addgene: pYPQ-Cas9-Act3.0 (no. 178954), pYPQ-CBE-Cas9n-Act3.0 (no. 178955), pYPQ-ABE-Cas9n-Act3.0 (no. 178956), pYPQ-SpRY-Act3.0 (no. 178957), pYPQ-CBE-SpRYn-Act3.0 (no. 178958), pYPQ-ABE-SpRYn-Act3.0 (no. 178959), pYPQ-132-tRNA (no. 179211), pYPQ-133-tRNA (no. 179212), pYPQ-134-tRNA (no. 179213) and pYPQ-134B (no. 179216). The sequence data of targeted genes can be found from The Arabidopsis Information Resource (https://www.Arabidopsis.org/), Rice Genome Annotation Project (http://rice.uga.edu/) Solanaceae Genomics Network (https://solgenomics.net/) or Phytozome (https://phytozome-next.jgi.doe.gov/) using their locus identifiers as follows: OsBBM1 (LOC_Os11g19060), OsGW2 (LOC_Os02g14720), OsGN1a (LOC_Os01g10110), OsALS (LOC_Os02g30630), OsEPSPS (LOC_Os06g04280), OsYSA (LOC_Os03g40020), OsMAPK5 (LOC_Os03g17700), AtFT (AT1G65480), AtAP1 (AT1G69120), AtPYL1 (AT5G46790), AtALS (AT3G48560), AtACC2 (AT1G36180), PtWUS (Potri.005G114700), PtWOX11 (Potri.013G066900), PtARK1 (Potri.011G011100), Pt4CL1 (Potri.001G036900), SFT (Solyc03g063100) and SolyA7 (Solyc01g010970). The NGS data have been deposited to National Center for Biotechnology Information (accession code PRJNA779678; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA779678).
References
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Zhang, Y., Malzahn, A. A., Sretenovic, S. & Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794 (2019).
Gao, C. Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635 (2021).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Molla, K. A., Sretenovic, S., Bansal, K. C. & Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 7, 1166–1187 (2021).
Grunewald, J. et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 38, 861–864 (2020).
Zhang, X. et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 38, 856–860 (2020).
Sakata, R. C. et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 38, 865–869 (2020).
Li, C. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–882 (2020).
Li, C. et al. SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol. 21, 141 (2020).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Lowder, L. G. et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985 (2015).
Lowder, L. G. et al. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-act systems. Mol. Plant 11, 245–256 (2018).
Pan, C., Sretenovic, S. & Qi, Y. CRISPR/dCas-mediated transcriptional and epigenetic regulation in plants. Curr. Opin. Plant Biol. 60, 101980 (2021).
Esvelt, K. M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Boettcher, M. et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat. Biotechnol. 36, 170–178 (2018).
Bai, M. et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731 (2020).
Liu, H. J. et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 32, 1397–1413 (2020).
Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 12, 1051–1054 (2015).
Singh, D. et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. USA 115, 5444–5449 (2018).
Breinig, M. et al. Multiplexed orthogonal genome editing and transcriptional activation by Cas12a. Nat. Methods 16, 51–54 (2019).
Pan, C. et al. CRISPR-Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 7, 942–953 (2021).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Ren, Q. et al. PAM-less plant genome editing using a CRISPR-SpRY toolbox. Nat. Plants 7, 25–33 (2021).
Ren, J. et al. Expanding the scope of genome editing with SpG and SpRY variants in rice. Sci. China Life Sci. 64, 1784–1787 (2021).
Xu, Z. et al. SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol. 22, 6 (2021).
Ren, Q. et al. Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol. J. 19, 2052–2068 https://doi.org/10.1111/pbi.13635 (2021).
Li, G., Sretenovic, S., Eisenstein, E., Coleman, G. & Qi, Y. Highly efficient C‐to‐T and A‐to‐G base editing in a Populus hybrid. Plant Biotechnol. J. https://doi.org/10.1111/pbi.13581 (2021).
Randall, L. B. et al. Genome- and transcriptome-wide off-target analyses of an improved cytosine base editor. Plant Physiol. https://doi.org/10.1093/plphys/kiab264 (2021).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Lapinaite, A. et al. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science 369, 566–571 (2020).
Altpeter, F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016).
Lowe, K. et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell https://doi.org/10.1105/tpc.16.00124 (2016).
Maher, M. F. et al. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89 (2020).
Debernardi, J. M. et al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279 (2020).
Li, J. et al. The WUSCHELa (PtoWUSa) is involved in developmental plasticity of adventitious root in poplar. Genes 11, https://doi.org/10.3390/genes11020176 (2020).
Liu, B. et al. PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar. Plant Mol. Biol. 98, 389–406 (2018).
Liu, X., Zhang, Z., Bian, W., Duan, A. & Zhang, H. Enhancing the expression of ARK1 genes in poplar leads to multiple branches and transcriptomic changes. R. Soc. Open Sci. 7, 201201 (2020).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019).
Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018).
Wang, Z. P. et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015).
Khanday, I., Santos-Medellín, C. & Sundaresan, V. Rice embryogenic trigger BABY BOOM1 promotes somatic embryogenesis by upregulation of auxin biosynthesis genes. bioRxiv https://doi.org/10.1101/2020.08.24.265025 (2020).
Meng, X. et al. Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant https://doi.org/10.1016/j.molp.2017.06.006 (2017).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Leple, J. C., Brasileiro, A. C., Michel, M. F., Delmotte, F. & Jouanin, L. Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep. 11, 137–141 (1992).
Liu, Q. et al. Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7 (2019).
Stewart, C. N. Jr. & Via, L. E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–750 (1993).
You, Q. et al. CRISPRMatch: an automatic calculation and visualization tool for high-throughput CRISPR genome-editing data analysis. Int. J. Biol. Sci. 14, 858–862 (2018).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Acknowledgements
This work was supported by the NSF Plant Genome Research Program grants (award nos. IOS-1758745 and IOS-2029889), the USDA-NIFA Biotechnology Risk Assessment grant (award no. 2020-33522-32274), the USDA-AFRI Agricultural Innovations Through Gene Editing Program (award no. 2021-67013-34554) and Maryland Innovation Initiative Funding (award no. 1120-012_2) to Y.Q. and a USDA-NIFA grant (award no. 2019-67013-29197) and a USDA McIntire-Stennis project (award no. MD-PSLA-20006) to G.C. A.M. was supported by NRT-INFEWS: UMD Global STEWARDS (STEM Training at the Nexus of Energy, Water Reuse and Food Systems) that was awarded to the University of Maryland School of Public Health by the NSF National Research Traineeship Program (award no. 1828910). S.S. is a fellow of Foundation for Food and Agriculture Research.
Author information
Authors and Affiliations
Contributions
Y.Q. and C.P. conceived the project and designed the experiments. C.P., B.L. and S.S. generated all the constructs. C.P. and Y.C. performed all experiments of rice and tomato. A.M. generated stable transgenic Arabidopsis. C.P. conducted molecular and phenotype analysis on these lines. G.L. and G.C. generated stable poplar plants and conducted the analysis. F.G. provided help during stable transformation of rice. Y.Q. and C.P. wrote the paper with input from other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.Q. and C.P. are inventors on a US Patent Application that has been filed on the CRISPR-Combo system in this study. Y.Q. is a consultant for Inari Agriculture and CTC Genomics. All other authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Jian-feng Li, Sadiye Hayta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CRISPR-Cas9-Act3.0 enables genome editing and gene activation in rice protoplasts programmed by guide RNA scaffolds and protospacer length.
a, Restriction fragment length polymorphism (RFLP) analysis of CRISPR-Cas9-mediated genome editing with different protospacer length (14-nt to 20-nt) in gR1.0. The Cas9 and single guide (sgRNA) scaffold gR1.0 were driven by ZmUbi and OsU3 promoters, respectively. Three biological replicates were performed for each target site. Representative gel images are shown. CTRL indicates wild-type samples. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). NLS, nuclear localization signal. b, Comparison of the editing efficiency between Cas9 and Cas9-Act3.0 with both 20- and 15-nt protospacers. The Cas9-Act3.0 system here consists of a catalytically active Cas9 nuclease and MS2 bacteriophage coat protein (MCP)-SunTag-activator complex, and a gR1.0. Three biological replicates were performed for each target site. Representative gel images are shown. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). P values were obtained using the two-tailed Student’s t-test. c, Cas9-Act3.0-induced gene activation with different protospacer length in gR2.0. The Cas9-Act3.0 system here consists of a catalytically active Cas9 nuclease and MS2 bacteriophage coat protein (MCP)-SunTag-activator complex, and a sgRNA2.0 (gR2.0) scaffold. T-DNA vectors without sgRNAs served as the negative control (CTRL). OsTubulin was selected as the endogenous control gene. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments).
Extended Data Fig. 2 Cleavage activity and deletion profiles of Cas9-Act3.0 in both rice and tomato protoplasts.
a,b, Comparison of the deletion position and size between Cas9 and Cas9-Act3.0 in rice (a) and tomato (b) protoplasts based on NGS data. Frequencies of deletion position were calculated using the number of reads with deletions at designated nucleotide position divided by the total reads with deletions. PAM is highlighted in red underline and protospacer sequence is highlighted in black underline. Frequencies of deletion size were calculated using the number of reads with designated size deletion divided by the total reads with deletions. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). c, Cleavage activity of Cas9-Act3.0 with 15-nt and 20-nt protospacers at the OsBBM1 and SFT promoters. One 15-nt and 20-nt protospacer for each of OsBBM1 and SFT was separately cloned into the gR2.0 scaffold. One 20-nt protospacer for each of OsGW2, OsGN1a, and SolyA7 was separately cloned into the gR1.0 scaffold. T-DNA vectors without sgRNAs served as the negative control (CTRL). Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). The different letters indicate significantly different mean values at P < 0.05 (one-way analysis of variance (ANOVA) with post-hoc Tukey test).
Extended Data Fig. 3 Development and characterization of the PAM-less SpRY-Act3.0 system.
a,b, Determination of SpRY-Act3.0-based-simultaneous gene activation (a) and targeted mutagenesis (b) in rice protoplasts at both NGG and NGC protospacer adjacent motifs (PAMs). One 15-nt protospacer of OsBBM1 each at both NGG and NGC PAMs was separately cloned into gR2.0 for SpRY-Act3.0-mediated gene activation. One 20-nt protospacer each for OsGW2 and OsGN1a at both NGG and NGC PAMs was separately cloned into gR1.0 for SpRY-Act3.0-mediated genome editing. The indel mutation assays were analysed by NGS. The dSpRY-Act3.0 activation system and SpRY nuclease were selected as references for comparing SpRY-Act3.0-mediated simultaneous gene activation and indel mutation, respectively. T-DNA vectors without sgRNAs served as the negative control (CTRL). OsTubulin was selected as the endogenous control gene. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). P values were obtained using the two-tailed Student’s t-test. *P < 0.05, **P < 0.01. c, Deletion position analysis of both SpRY and SpRY-Act3.0 systems in rice protoplasts based on NGS data. Deletion frequencies were calculated using the number of reads with deletions at designated nucleotide position divided by the total reads with deletions. PAM is highlighted in red underline and protospacer sequence is highlighted in black underline. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). d, Deletion size analysis of both SpRY and SpRY-Act3.0 systems in rice protoplasts. Deletion frequencies were calculated using the number of reads with designated size deletion divided by the total reads with deletions. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments).
Extended Data Fig. 4 Editing activity and window analysis of CBE-Cas9n-Act3.0 and ABE-Cas9n-Act3.0 systems in rice and tomato protoplasts.
a,b, Editing window analysis of CBE-Cas9n and CBE-Cas9n-Act3.0 systems in rice (a) and tomato (b) protoplasts based on NGS data. The C to T conversion efficiencies were analysed by the CRISPR RGEN tools. ‘Cn’ indicates the position of target C in the protospacer. Error bar represents the mean ± s.d. (n = 3 independent experiments). c, Editing window analysis of ABE-Cas9n and ABE-Cas9n-Act3.0 base editors in rice protoplasts. The A to G conversion efficiencies were analysed by the CRISPR RGEN tools. ‘An’ indicates the position of target A in the protospacer. Error bar represents the mean ± s.d. (n = 3 independent experiments). d, Editing activity of CBE-Cas9n-Act3.0 and CBE-Cas9n-Act3.0 systems with 15-nt and 20-nt protospacers at the OsBBM1 and SFT promoters. One 15-nt and 20-nt protospacer for each of OsBBM1 and SFT was separately cloned into the gR2.0 scaffold. One 20-nt protospacer for each of OsALS, OsEPSPS, and SolyA7 was separately cloned into the gR1.0 scaffold. T-DNA vectors without sgRNAs served as the negative control (CTRL). Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). The different letters indicate significantly different mean values at P < 0.05 (one-way analysis of variance (ANOVA) with post-hoc Tukey test).
Extended Data Fig. 5 Characterization of the CBE-SpRYn-Act3.0 and ABE-SpRYn-Act3.0 systems.
a,b, Determination of CBE-SpRYn-Act3.0 and ABE-SpRYn-Act3.0-based-simultaneous gene activation (a) and base editing (b) efficiency in rice protoplasts. One 15-nt protospacer of OsBBM1 was cloned into gR2.0 for CBE/ABE-SpRYn-Act3.0-mediated gene activation. One 20-nt protospacer for both OsALS and OsEPSPS each was separately cloned into gR1.0 for CBE/ABE-SpRYn-Act3.0-mediated base editing. A indicates CBE/ABE-SpRYn-Act3.0-mediated target gene activation with only gR2.0 scaffold. A + BE indicates CBE/ABE-SpRYn-Act3.0-mediated simultaneous gene activation and base editing with both gR1.0 and gR2.0 scaffolds. To compare base editing efficiency, CBE-SpRYn and ABE-SpRYn were selected as references in CBE-SpRYn-Act3.0 and ABE-SpRYn-Act3.0-mediated A + BE assays, respectively. T-DNA vectors without sgRNAs served as the negative control (CTRL). OsTubulin was selected as the endogenous control gene. Each dot represents an individual biological replicate. Error bar represents the mean ± s.d. (n = 3 independent experiments). P values were obtained using the two-tailed Student’s t-test. *P < 0.05, **P < 0.01. c,d, Editing window analysis of CBE-SpRYn-Act3.0 (c) and ABE-SpRYn-Act3.0 (d) base editors in rice protoplasts. The C to T and A to G conversion efficiencies were analysed by the CRISPR RGEN tools. ‘Cn’ and ‘An’ indicate the position of target C and A in the protospacer, respectively. Error bar represents the mean ± s.d. (n = 3 independent experiments).
Extended Data Fig. 6 Mutation and genotype analysis of CBE-Cas9n-Act3.0-mediated T2 standard flowering plants (transgene-free).
a, Mutation analysis of T2 T-DNA free standard flowering populations for the CBE-Cas9n-Act3.0 system. The base editing frequencies of examined transgene-free plants were determined by NGS. Each dot indicates an individual plant. Error bar represents the mean ± s.d. (n = 29, 24, 37, 38 and 32 independent plants for #5, #14, #15, #16 and #17, respectively). b, Representative genotypes of atalsatacc2, atalsAtACC2 and AtALSatacc2 plants in T2 T-DNA free standard flowering populations. Three kinds of genotypes (atalsatacc2, atalsAtACC2, and AtALSatacc2) are identified in CBE-Cas9n-Act3.0-mediated standard flowering (transgene-free) populations. The PAM is highlighted in yellow. The DNA bases C in the protospacer sequence are highlighted in red, and the other DNA bases of the protospacer are highlighted in green. The symbols above the protospacer sequence indicate amino acids. The numbers below the protospacer sequence indicate the percentage of DNA base T or C in total reads. The base editing efficiencies were analysed by the CRISPR RGEN tools.
Extended Data Fig. 7 Undetectable off-target genome-editing activity by Cas9-Act3.0 and CBE-Cas9n-Act3.0 at the AtFT promoter with 15-nt protospacers.
a, Identification of Cas9-Act3.0-mediated potential indel mutations at two AtFT target sites in both T2 extra-early flowering and T-DNA free standard flowering plants. Three and two independent lines were selected for extra-early flowering and T-DNA free standard flowering groups, respectively. Approximately 15 to 23 individual plants were examined for each line. n/a, no indel mutation was detected. 1d1/1,527, one 1-bp deletion event detected in a total of 1,527 reads. 1d1/838, one 1-bp deletion event was detected in a total of 838 reads. The indel mutations were analysed by CRISPResso2. b, Identification of CBE-Cas9n-Act3.0-mediated potential base editing at AtFT-sgRNA1 target site in both T2 extra-early flowering and T-DNA free standard flowering plants. Three and two independent lines were selected for extra-early flowering and T-DNA free standard flowering groups, respectively. CTRL represents wild-type plants. Approximately 15 to 24 individual plants were examined for each line. The PAM of AtFT-sgRNA1 is highlighted in red and DNA bases C in the protospacer sequence are highlighted in green. The base editing efficiencies were analysed by the CRISPR RGEN tools. Each dot represents an individual plant. Error bar represents the mean ± s.d. (n = 15 independent plants for CTRL, #5, #16 and #17, n = 24 independent plants for T-DNA free lines #5 and #17). Note the AtFT-sgRNA2 doesn’t contain any DNA base C and hence was not analysed for C to T base editing.
Extended Data Fig. 8 Cas9-Act3.0 promotes de novo callus and root organogenesis from petiole and stem cuttings by activation of PtWUS in poplar.
a, Cas9-Act3.0 promotes root initiation by activation of PtWUS in poplar. Two gR2.0 with 15-nt protospacers and one gR1.0 with 20-nt protospacer were used for PtWUS and Pt4CL1, respectively. All transgenic plants were grown in the root-induction medium (RIM) with hygromycin selection. For days to root, error bar represents the mean ± s.d. (n = 11 and 13 independent plants for Cas9 and Cas9-Act3.0, respectively). For rooting rate, error bar represents the mean ± s.d. (n = 3 biological replicates). The root initiation rate was evaluated on the 7th day after transgenic shoots were transferred to RIM. b, Representative genotypes of Cas9-Act3.0 and Cas9-mediated T0 Pt4CL1 mutants. The red dash indicates a nucleotide deletion. The bold DNA bases indicate insertion. PAM, protospacer adjacent motif. c, Cas9-Act3.0 promotes de novo callus formation from petiole cuttings and leaf-disc regeneration by activation of PtWUS. Petiole cuttings and leaf discs from CTRL and PtWUS-activation poplar plants were cultured on callus induction medium (CIM) for two weeks. Representative PtWUS-activation lines including #2, #4, and #17 are shown. d, The Cas9-Act3.0 promotes shoot growth of stem cuttings by activation of PtWUS in poplar. Four stem cuttings from each of CTRL and PtWUS-activation line (#4) were cultured in one magenta box with root-induction medium (RIM). e, Cas9-Act3.0 promotes de novo root initiation of stem cuttings by activation of PtWUS in poplar. Five to seven stem cuttings were cultured in one magenta box with the root-induction medium. Red dash cycles indicate successful root initiation and white dash cycles indicate failed root initiation. f,h, Representative shoot (f) and root (h) phenotype of CTRL and PtWUS-activation poplar line (#2). Plants were grown in soil under the same cultivation conditions for six weeks. Error bar represents the mean ± s.d. (n = 4 independent plants). g, Leaf area analysis of CTRL and PtWUS-activation poplar lines (#2 and #4). Error bar represents the mean ± s.d. (n = 4 independent plants). P values in a, f-h were obtained using the two-tailed Student’s t-test. **P < 0.01.
Extended Data Fig. 9 Cas9-Act3.0 promotes de novo callus and root organogenesis of petiole and shoot regenerated from PtWOX11-activation poplar.
a, Representative genotypes of Cas9-Act3.0 and Cas9-mediated T0 Pt4CL1 mutants. The four DNA bases A, T, C, G are marked in different colours. The red dash indicates a nucleotide deletion. The bold DNA bases indicate insertion. PAM, protospacer adjacent motif. b, Cas9-Act3.0 promotes de novo callus formation from petiole cuttings by activation of PtWOX11. Petiole cuttings from CTRL and PtWOX11-acitvation plants were cultured on callus induction medium (CIM) for three weeks. Representative petioles of PtWOX11-acitvation lines including #2, #3, and #6 are shown. c, The Cas9-Act3.0 system promotes root initiation of shoots regenerated from the PtWOX11-activation poplar. Four regenerated shoots from each of CTRL and PtWOX11-acitvation plants were cultured in one magenta box with root-induction medium (RIM) for seven days. Representative root and shoot of CTRL and PtWOX11-acitvation lines are shown. Error bar represents the mean ± s.d. (n = 4 independent plants). d, Representative shoots of CTRL and PtWOX11-acitvation lines. Plants were grown in soil under the same cultivation conditions for six weeks. Three or four independent plants were examined for both CTRL and PtWOX11-acitvation lines. e, Leaf area analysis of CTRL and PtWOX11-activation poplar lines. Each dot indicates the average area of all leaves for an individual plant. Error bar represents the mean ± s.d. (n = 4 independent plants). P values in c,e were obtained using the two-tailed Student’s t-test. *P < 0.05, **P < 0.01.
Extended Data Fig. 10 Cas9-Act3.0 promotes callus regeneration and enriches heritable targeted mutations by activation of OsBBM1 in rice.
a,b, Representative images of Cas9-Act3.0-induced shoot regeneration by activation of OsBBM1 at four (a) and seven (b) weeks. The calli of both vectors were grown on regeneration I (Reg I) medium for seven weeks by subculturing every two weeks. c, Zygosity analysis of Cas9-Act3.0-mediated T0 lines at the OsGN1a target site. A total of 10 and 16 individual transgenic plants were examined for Cas9-Act3.0-based GE and A+GE vectors, respectively. GE, genome editing. A+GE, simultaneous gene activation and editing. d,j, Representative images of Cas9-Act3.0-induced rice plants from hormone-containing and hormone-free (HF) mediums. e, Representative images of Cas9-Act3.0-mediated callus induction and regeneration by activating OsBMM1 in a HF manner. RSM, rice selection medium. All calli were subcultured with HF fresh medium every two weeks. f, Efficiency of Cas9-Act3.0-callus induction and regeneration in a HF manner. All calli were cultured under the same conditions. g, Number of regenerated transgenic plants per Cas9-Act3.0-mediated callus in a HF manner. Each dot indicates the seedling number generated from one transgenic callus. Error bar represents the mean ± s.d. (n = 10 independent calli). h, Zygosity analysis of Cas9-Act3.0-mediated T0 OsGW2OsGN1a mutants in a HF manner. The frequencies of each zygotic type are shown as percentages. A total of 14 individual transgenic plants were examined using NGS. i, Representative genotypes of Cas9-Act3.0-mediated T0 OsGW2OsGN1a mutants in a HF manner. The red dash indicates a nucleotide deletion. The black dash indicates a blank space. The bold DNA bases indicate insertion. PAM, protospacer adjacent motif. k, Analysis of the cleavage activity of Cas9-Act3.0 at the target sites of OsBBM1 promoter in rice plants. Both tgR1 and tgR2 of a 15-nt protospacer were used for OsBBM1 activation. Error bar represents the mean ± s.d. (n = 5 and 18 independent plants for CTRL and A+GE, respectively). P values were obtained using the two-tailed Student’s t-test. l, Determination of OsBBM1 activation level in Cas9-Act3.0-mediated HF edited plants using qRT–PCR. Leaf tissue was sampled for total RNA extraction. CTRL represents the mixed sample from three individual plants. Error bar represents the mean ± s.d. (n = 3 technical replicates).
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–7 and Tables 1–3.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, C., Li, G., Malzahn, A.A. et al. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 8, 513–525 (2022). https://doi.org/10.1038/s41477-022-01151-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01151-9
This article is cited by
-
Precise fine-turning of GhTFL1 by base editing tools defines ideal cotton plant architecture
Genome Biology (2024)
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
CRISPR/Cas-mediated germplasm improvement and new strategies for crop protection
Crop Health (2024)
-
Breeding rice for yield improvement through CRISPR/Cas9 genome editing method: current technologies and examples
Physiology and Molecular Biology of Plants (2024)
-
Efficient and versatile multiplex prime editing in hexaploid wheat
Genome Biology (2023)