Abstract
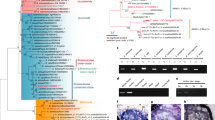
In flowering plants, different lineages have independently transitioned from the ancestral hermaphroditic state into and out of various sexual systems1. Polyploidizations are often associated with this plasticity in sexual systems2,3. Persimmons (the genus Diospyros) have evolved dioecy via lineage-specific palaeoploidizations. More recently, hexaploid D. kaki has established monoecy and also exhibits reversions from male to hermaphrodite flowers in response to natural environmental signals (natural hermaphroditism, NH), or to artificial cytokinin treatment (artificial hermaphroditism, AH). We sought to identify the molecular pathways underlying these polyploid-specific reversions to hermaphroditism. Co-expression network analyses identified regulatory pathways specific to NH or AH transitions. Surprisingly, the two pathways appeared to be antagonistic, with abscisic acid and cytokinin signalling for NH and AH, respectively. Among the genes common to both pathways leading to hermaphroditic flowers, we identified a small-Myb RADIALIS-like gene, named DkRAD, which is specifically activated in hexaploid D. kaki. Consistently, ectopic overexpression of DkRAD in two model plants resulted in hypergrowth of the gynoecium. These results suggest that production of hermaphrodite flowers via polyploidization depends on DkRAD activation, which is not associated with a loss-of-function within the existing sex determination pathway, but rather represents a new path to (or reinvention of) hermaphroditism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. All sequence data generated in the context of this manuscript have been deposited in the appropriate DNA Database of Japan: Illumina reads for mRNA-seq in the Short Read Archives (SRA) database (SRA Submission ID: DRA013154, Run IDs: DRR332477–332618).
References
Käfer, J., Marais, G. A. & Pannell, J. R. On the rarity of dioecy in flowering plants. Mol. Ecol. 26, 1225–1241 (2017).
Ashman, T. L., Kwok, A. & Husband, B. C. Revisiting the dioecy-polyploidy association: alternate pathways and research opportunities. Cytogenet. Genome Res. 140, 241–255 (2013).
Goldberg, E. E. Macroevolutionary synthesis of flowering plant sexual systems. Evolution 71, 898–912 (2017).
Renner, S. S. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101, 1588–1596 (2014).
Heilbuth, J. C. Lower species richness in dioecious clades. Am. Nat. 156, 221–241 (2000).
Charlesworth, B. & Charlesworth, D. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997 (1978).
Renner, S. S. & Ricklefs, R. E. Dioecy and its correlates in the flowering plants. Am. J. Bot. 82, 596–606 (1995).
Barrett, S. C. The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274–284 (2002).
Liu, Z. et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427, 348–352 (2004).
Wang, J. et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl Acad. Sci. USA 109, 13710–13715 (2012).
Kazama, Y. et al. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 6, 18917 (2016).
Krasovec, M., Chester, M., Ridout, K. & Filatov, D. A. The mutation rate and the age of the sex chromosomes in Silene latifolia. Curr. Biol. 28, 1832–1838 (2018).
Torres, M. F., Mohamoud, Y. A., Younuskunju, S., Suhre, K. & Malek, J. A. Evidence of recombination suppression blocks on the Y chromosome of date palm (Phoenix dactylifera). Front. Plant Sci. 12, 634901 (2021).
Akagi, T., Henry, I. M., Tao, R. & Comai, L. A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650 (2014).
Akagi, T. et al. The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 16, e1008566 (2020).
Harkess, A. et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 8, 1279 (2017).
Harkess, A. et al. Sex determination by two Y-linked genes in garden asparagus. Plant Cell 32, 1790–1796 (2020).
Murase, K. et al. MYB transcription factor gene involved in sex determination in Asparagus officinalis. Genes Cells 22, 115–123 (2017).
Tsugama, D. et al. A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis. Sci. Rep. 7, 41497 (2017).
Akagi, T. et al. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30, 780–795 (2018).
Akagi, T. et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 5, 801–809 (2019).
Tennessen, J. A. et al. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 16, e2006062 (2018).
Torres, M. et al. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 9, 3969 (2018).
Massonnet, M. et al. The genetic basis of sex determination in grapes. Nat. Commun. 11, 2902 (2020).
Müller, N. A. et al. A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 6, 630–637 (2020).
Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005).
Akagi, T., Henry, I. M., Kawai, T., Comai, L. & Tao, R. Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28, 2905–2915 (2016).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Wendel, J. F. Genome evolution in polyploids. Plant Mol. Evol. 42, 225–249 (2000).
Osborn, T. C. et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147 (2003).
Yang, H. W., Akagi, T., Kawakatsu, T. & Tao, R. Gene networks orchestrated by MeGI: a single-factor mechanism underlying sex determination in persimmon. Plant J. 98, 97–111 (2019).
Bawa, K. S. Evolution of dioecy in flowering plants. Annu. Rev. Ecol. Syst. 11, 15–39 (1980).
Akagi, T., Kawai, T. & Tao, R. A male determinant gene in diploid dioecious Diospyros, OGI, is required for male flower production in monoecious individuals of Oriental persimmon (D. kaki). Sci. Hort. 213, 243–251 (2016).
Spongberg, S. A. Notes on persimmons, kakis, date plums, and chapotes. Arnoldia 39, 290–309 (1979).
Wallnöfer, B. The biology and systematics of Ebenaceae: a review. Ann. Nat. Mus. Wien B 103, 485–512 (2001).
Yonemori, K., Yomo, Y. & Sugiura, A. Sexuality in Japanese persimmons. 2. Induction of sex conversion in male flower by cytokinin treatment. J. Jpn. Soc. Hort. Sci. 59, 230–231 (1990).
Yonemori, K. et al. Sequence analyses of the ITS regions and the matK gene for determining phylogenetic relationships of Diospyros kaki (persimmon) with other wild Diospyros (Ebenaceae) species. Tree Genet. Genomes 4, 149–158 (2008).
Akagi, T., Tao, R., Tsujimoto, T., Kono, A. & Yonemori, K. Fine genotyping of a highly polymorphic ASTRINGENCY-linked locus reveals variable hexasomic inheritance in persimmon (Diospyros kaki Thunb.) cultivars. Tree Genet. Genomes 8, 195–204 (2012).
Corley, S. B., Carpenter, R., Copsey, L. & Coen, E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl Acad. Sci. USA 102, 5068–5073 (2005).
Costa, M. M. R., Fox, S., Hanna, A. I., Baxter, C. & Coen, E. Evolution of regulatory interactions controlling floral asymmetry. Development 132, 5093–5101 (2005).
Lucibelli, F., Valoroso, M. C. & Aceto, S. Radial or bilateral? The molecular basis of floral symmetry. Genes 11, 395 (2020).
Hamaguchi, A. et al. A small subfamily of Arabidopsis RADIALIS-LIKE SANT/MYB genes: a link to HOOKLESS1-mediated signal transduction during early morphogenesis. Biosci. Biotechnol. Biochem. 72, 2687–2696 (2008).
Akagi, T. & Charlesworth, D. Pleiotropic effects of sex-determining genes in the evolution of dioecy in two plant species. Proc. R. Soc. B 286, 20191805 (2019).
Rawat, R. et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl Acad. Sci. USA 106, 16883–16888 (2009).
Rawat, R. et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7, e1001350 (2011).
Li, S. & Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913 (2013).
Yang, Y. et al. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 39, e101928 (2020).
Martínez-Fernández, I. et al. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front. Plant Sci. 5, 210 (2014).
Wang, L. et al. Arabidopsis ADF1 regulated by MYB73 and is involved in response to salt stress via affecting actin filaments organization. Plant Cell Physiol. 62, 1387–1395 (2021).
Marsch-Martínez, N. & de Folter, S. Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 29, 104–114 (2016).
Roldan, M. V. G. et al. Integrative genome-wide analysis reveals the role of WIP proteins in inhibition of growth and development. Commun. Biol. 3, 239 (2020).
Picq, S. et al. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol. 14, 229 (2014).
VanBuren, R. et al. Extremely low nucleotide diversity in the X-linked region of papaya caused by a strong selective sweep. Genome Biol. 17, 230 (2016).
VanBuren, R. et al. Origin and domestication of papaya Yh chromosome. Genome Res. 25, 524–533 (2015).
Van de Peer, Y., Ashman, T. L., Soltis, P. S. & Soltis, D. E. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33, 11–26 (2021).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Gupta, A. et al. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci. Rep. 7, 4017 (2017).
Shimada, T. L., Shimada, T. & Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010).
De Rybel, B. et al. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 156, 1292–1299 (2011).
Horsch, R. B., Rogers, S. G. & Fraley, R. T. Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437 (1985).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Acknowledgements
We thank T. Saito, N. Onoue and R. Matsuzaki (Grape and Persimmon Research Station, NIFTS, Japan) for some plant materials; and Ho-Wen Yang (University of Illinois) for experimental support. This work was supported by PRESTO from Japan Science and Technology Agency (JST) (grant nos. JPMJPR20D1 to T.A.); Grant-in-Aid for Scientific Research on Innovative Areas from JSPS (grant nos. 19H04862 to T.A. and 20H05391 to Y.I.); the Joint Usage/Research Center, Institute of Plant Science and Resources, Okayama University (to Y.I. and T.A.); and Grant-in-Aid for JSPS Fellows (grant nos. 19J23361 to K.M.).
Author information
Authors and Affiliations
Contributions
T.A. conceived the study and designed the experiments. K.M., Y.I., T.M., T.K. and T.A. conducted the experiments. K.M. and T.A. analysed the data. Y.I., T.K., R.T., Y.K., K.U., I.M.H. and T.A. contributed to plant resources and facilities. K.M., Y.I., T.K., I.M.H. and T.A. drafted the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Mathias Scharmann, Roberta Bergero and Yusuke Kazama for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Morphological characterization of male, NH, and AH flowers at various organ developmental stages.
a-l, Flower cross-sections stained with toluidine blue at stage 2 (a-c) and stage 3 (d-f) (see details in Supplementary Fig. 1). Dissected flower (g-i) and pollen fertility (j-l) at the mature stage. Ovu, ovule; Pe, Petal; Pt, Pollen tube; Sp, Sepal. Scale bars: 0.5 mm for stages 2 and 3, 1.0 mm for the mature stage, and 0.2 mm for pollen germination. o, MeGI expressions in NH and AH flowers were not upregulated from male flowers (P = 0.82-0.97), and significantly lower than female flowers during the organ developmental processes (P < 0.005 at stage 2 and P < 8.0e-4 at stage 3 with a one-sided Student’s t-test; n = total 61 and 72 samples at stage 2 and 3, respectively, see detail in Supplementary Table 1). *** indicate P < 0.005 for statistical significance. P = 0.005, 0.004, 0.003 for M, NH, AH in comparison to F at stage 2, respectively. P = 8.0e-4, 5.0e-4, and 5.0e-4 for M, NH, AH in comparison to F at stage 3, respectively.
Extended Data Fig. 2 Clustering of differentially expressed genes (DEGs) between NH/AH and male flowers in stage 2.
a, Number of differentially expressed genes (DEGs) between NH/AH and male flowers in stage 2 (FDR < 0.1, bias > 1.2-fold). b, Expression patterns of NH and AH upregulated DEGs in stage 2 (n = 507 and 703, respectively). NH upregulated genes were overall downregulated in AH. c, Clustering of the 5,163 DEGs revealed 14 clusters. d, The heatmap shows the distinct expression patterns of the four flower types in stage 2. e, Expression patterns of the four flower types in each of the 14 clusters. NH-specific, AH-specific, and NH/AH-shared upregulated clusters were defined as Clst 10, Clst 4, and Clst 5, respectively. *** indicate P < 2.2e-16 with two-sided Student’s t-test. The boxes spanned the interquartile range (25th to 75th percentiles), the center line indicated the median values, and the whiskers extended to 1.5x interquartile range.
Extended Data Fig. 3 Comparison of the expression patterns of all Clst5 genes and the three transcription factors present in Clst 5 (other than DkRAD), between hexaploid D. kaki and diploid D. lotus.
a, Expression clustering with the Clst 5 genes, in the control male, NH, male treated with 100 mg l−1 ABA, and male treated with 10 ppm CK, both in D. kaki and in D. lotus. Only DkRAD (RADIALIS, highlighted in pink) and glycosyl transferase-like genes exhibited statistically supported upregulation specific to the conversion into hermaphrodite flowers. b-d, All three transcription factors showed upregulation after the treatment with 100 mg l−1 ABA, not only in D. kaki but also in D. lotus. Their expression activation patterns in hexaploid D. kaki were much less correlated to the gynoecium restoration patterns (see Fig. 2d). P-values were obtained by comparing expression values using a one-sided t-test (n = total 69 samples, see details in Supplementary Table 1). The boxes spanned the interquartile range (25th to 75th percentiles), the center line indicated the median values, and the whiskers extended to 1.5x interquartile range.
Extended Data Fig. 4 Evolutionary tree of the RADIALIS family genes in angiosperms.
a, Phylogenetic tree of RADIALIS-like genes from representative eudicot species. We selected outgroup genes in the Arabidopsis thaliana genome (RAD1 and RAD3 clades), based on the previous study on clustering of RADIALIS-like genes (Gao et al. 2017). RADIALIS from Antirrhinum majus, which was originally identified as the RADIALIS gene, and DkRAD and DkRAD2 (from D. kaki) were indicated in green and red, respectively. They were nested into the same clade, the RAD2, with significant statistic support (bootstrap value = 0.93). b, Phylogenetic tree of RADIALIS-like genes within the RAD2 clade, with the genes from various angiosperms species (homology e-value < 1e−30), using CCA1-like genes from A. thaliana as the outgroup. Although this clade was thought to be basically monophyletic, each lineage underwent frequent duplications. DkRAD and DkRAD2 were also derived from a lineage-specific duplication. c, Expression patterns of the two RADIALIS-like genes, DkRAD and DkRAD2, in hexaploid D. kaki and diploid D.lotus. DkRAD expression was not detectable in diploid D. lotus, or in stage 3 in D.kaki, but it was expressed in stage 2 flowers of D. kaki. DkRAD2 exhibited AH-specific upregulation in D. kaki (as evidenced by the fact that belongs to Clst 4, Fig. 1f) and was expressed at some level in all samples examined, including diploid D. lotus.
Extended Data Fig. 5 Various side effects of the DkRAD over-expression Arabidopsis and Nicotiana tabacum transgenic lines.
a-c, Ectopic over-expression of DkRAD resulted in failure of self-fertilization in A. thaliana. Siliques developed normally after self-pollination in the control, while DkRAD ox. lines were self-sterile. Phenotype of the control line and DkRAD over-expression line #12 were shown. DSd, deficient seeds; Sd, seed; Si, silique; SS, sterile silique; Sty, Style. d, Number of seeds in control and the DkRAD over-expression lines (n = 3–9 biological replicates, see detail in Supplementary Table 8). P-values were detected in comparison to the control #1 using two-sided Student’s t-test. Data were expressed as mean values ± SD. f, Cross-pollination with control lines could produce fertile silique, suggesting that female organs are functional in the DkRAD transgenic lines (n = 3-4 biological replicates). Comparison of the fertile seed numbers between open-pollination and artificial self-pollination or reciprocal crossing with the control in DkRAD ox. #12. All the artificial self-pollination and reciprocal cross-pollination exhibited recovery of the seed fertility from the open-pollination, supporting that physical distance between anthers and stigma would be the cause of sterility in the DkRAD ox. lines. P-values were detected in comparison to DkRAD ox. #12 using two-sided Student’s t-test. Data were expressed as mean values ± SD. g, The control and DkRAD ox. lines showed no substantial differences in ovule development (immediately before the anthesis). Self-sterility of the DkRAD ox. lines could be due to the physical distance between stigma and anthers, caused by the hyper-growth of carpel. h, DkRAD ox. lines showed severe growth retardance, although their flowering timing was not substantially delayed from the control lines. i-j, Precocious flowering in the transgenic lines in comparison to the control. The DkRAD and DkRAD2 over-expression lines often formed flower buds even in selection media immediately after the regeneration in vitro (i). k, The DkRAD2 over-expression lines exhibit enlarged carpel before flowering in comparison to the control. Cp, carpel; FB, flower bud; St, stamen.
Extended Data Fig. 6 Phenotypic variations in carpel development, in the progeny of DkRAD-ox x pSyGI-SyGI lines.
a, Half of the stigma exhibited restoration of normal development. The area on the left (highlighted with a pink asterisk showed the pSyGl-SyGl phenotype with shrunken stigma. b, Weak restoration of stigma development. c-d, Carpel development was severely repressed in pSyGI-SyGl plants in flowering stage (Akagi et al. 2018) (c), while DkRAD overexpression restored the carpel development, often resulting in overgrowth (d). Cp, carpel; Pe, petal; Sg, stigma; St; stamen. e, Characterization of gynoecium development and carpel length in the 4 independent DkRAD ox. x pSyGI-SyGI lines.
Extended Data Fig. 7 Comparison of gene expression patterns in DkRAD overexpressing and control A. thaliana flower, and D. kaki leaves.
a, Detection of differentially expressed genes (DEGs) between the transgenic lines of DkRAD ox. and control developing flowers in A. thaliana (stages 8–10). Distribution of the expression patterns of the DEGs. The X and Y axes correspond to the normalized expression level (RPKM) and DkRAD ox./control flowers ratio, respectively. The 330 upregulated and 187 downregulated DEGs are highlighted in blue circles (FDR < 0.1, Supplementary Table 10). The DEGs with known auxin- and defense/stress-signals functions, or those involved in cell cycle or circadian rhythm regulation functions are indicated with pink circles. b, Visualization of the co-expression network exhibiting upregulation in DKRAD ox. Transcription factors are aligned at the top. c,d, Detection of DEGs between control and transiently DkRAD overexpressing D. kaki leaves. The 1,413 upregulated and 595 downregulated DEGs are shown in blue circles (FDR < 0.1) (c). Ven diagram of the DEGs identified in transiently DkRAD overexpressing flowers (blue circles), NH flowers (deep green circles) and AH flowers (orange circle) in persimmon, compared to control flowers (Extended Data Fig. 2a and Supplementary Table 12) (d). e, Hypothetical regulatory pathway to produce hermaphrodite flowers from male flowers in persimmon. Activation of DkRAD expression, which integrates NH and AH signals, would lead to upregulation of auxin and stress responsive signaling, potentially via MYB73 expression, resulting in promotion of gynoecium growth.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1–14.
Supplementary Table 1
Supplementary Tables 4, 6, 7, 10 and 12.
Rights and permissions
About this article
Cite this article
Masuda, K., Ikeda, Y., Matsuura, T. et al. Reinvention of hermaphroditism via activation of a RADIALIS-like gene in hexaploid persimmon. Nat. Plants 8, 217–224 (2022). https://doi.org/10.1038/s41477-022-01107-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01107-z
This article is cited by
-
Identification and analysis of miRNAs differentially expressed in male and female Trichosanthes kirilowii maxim
BMC Genomics (2023)
-
Genetic insights into the dissolution of dioecy in diploid persimmon Diospyros oleifera Cheng
BMC Plant Biology (2023)
-
Recurrent neo-sex chromosome evolution in kiwifruit
Nature Plants (2023)
-
Comparative transcriptome analysis reveals candidate genes related to the sex differentiation of Schisandra chinensis
Functional & Integrative Genomics (2023)
-
A symmetry gene restores femaleness
Nature Plants (2022)